Figure 2. The SNF2 family of proteins.
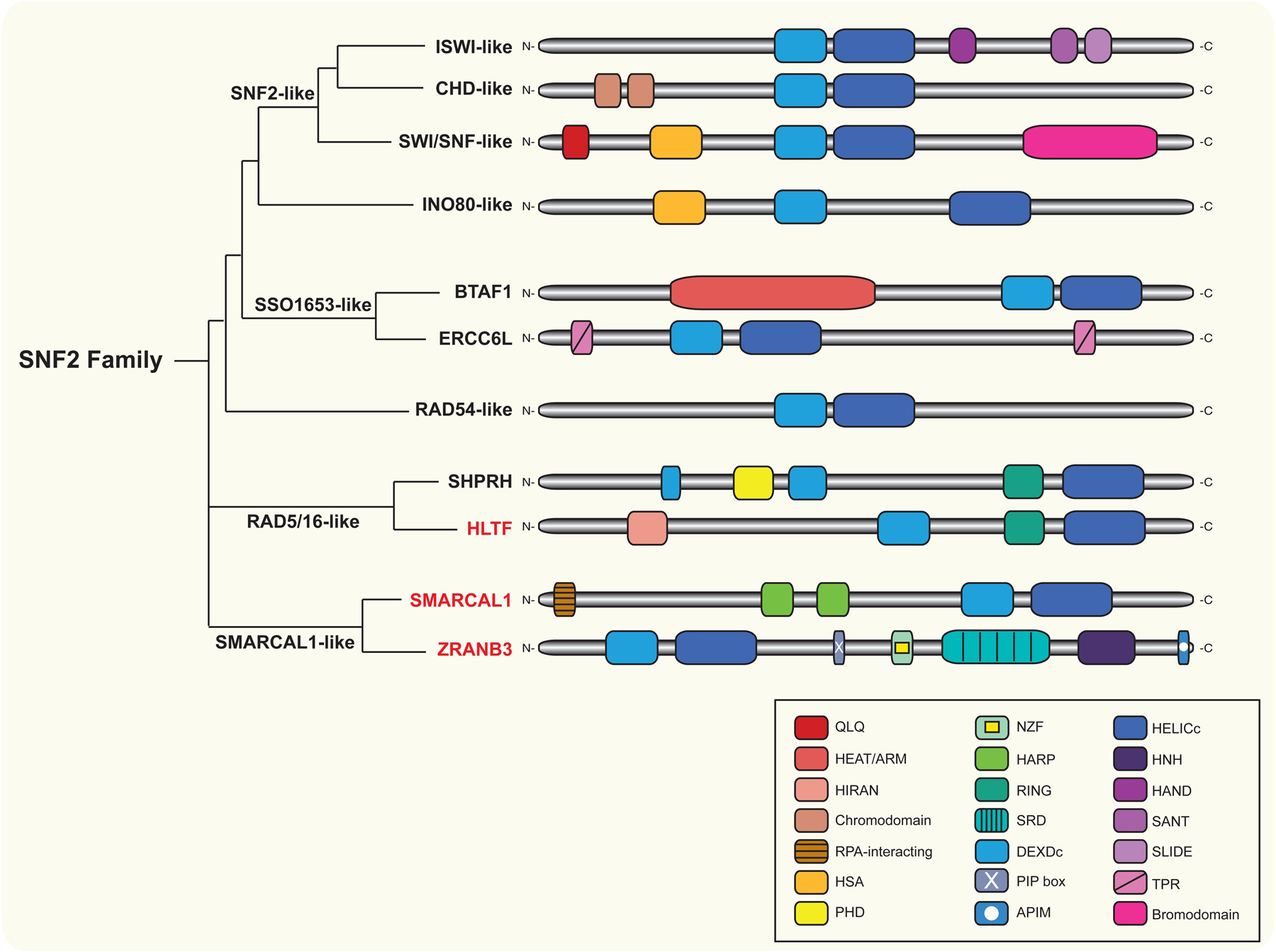
Depiction of SNF2-family group relationships, based on alignments of the DNA translocase domain (adapted from Flaus et al. [98]). Schematic representations (not to scale) of protein domains characteristic of each subfamily in humans are shown. All SNF2-family members contain a DNA translocase domain consisting of DEXDc and HELICc domains. The SNF2-like subfamily consists of ISWI-like, CHD-like and SWI/SNF-like groups. The ISWI-like group includes SMARCA1 (1054 aa) and SMARCA5 (1052 aa). The CHD-like group includes CHD1 (1710 aa), CHD2 (1828 aa), CHD3 (2000 aa), CHD4 (1912 aa), CHD5 (1954 aa), CHD6 (2715 aa), CHD7 (2997 aa), CHD8 (2581 aa), CHD9 (2897 aa) and CHD1L (897 aa). CHD3, CHD4 and CHD5 have N-terminal plant homeodomain (PHD) fingers not represented in the schematic, while CHD1L lacks the N-terminal chromodomains and contains a C-terminal macrodomain (not shown). The SWI/SNF-like group includes SMARCA2 (1590 aa) and SMARCA4 (1647 aa). HELLS (838 aa) is closely related to the SNF2-like subfamily, but it does not contain domains typical of the members of this subfamily (not shown). The INO80-like subfamily includes INO80 (1556 aa), SRCAP (3230 aa) and EP400 (3159 aa), which contain additional A/T hook and SANT domains, respectively, and SMARCAD1 (1026 aa), which lacks the HSA domain and contains two N-terminal coupling of ubiquitin conjugation to ER degradation (CUE) motifs not represented in the schematic. The SSO1653-like subfamily includes BTAF1 (1849 aa), which contains multiple HEAT/ARM repeats, and ERCC6L (1250 aa), which harbors two tetratricopeptide repeat (TPR) motifs and a PICH family domain (PFD) of unclear function not represented in the schematic. The RAD54-like subfamily includes RAD54 (747 aa) and ATRX (2492 aa), which contains an N-terminal ATRX-DNMT1-DNMT1L (ADD) domain not shown. The RAD5/16-like subfamily includes SHPRH (1683 aa), which contains a linker histone H1/H5 (H15) domain in addition to the domains represented in the figure, and HLTF (1009 aa). Finally, the SMARCAL1-like subfamily includes SMARCAL1 (954 aa) and ZRANB3 (1079 aa). SMARCAL1, ZRANB3 and HLTF (in red) are the only SNF2-family members that have been currently shown to regress stalled replication forks in vivo. HLTF contains a RING domain that mediates the interaction between HLTF and substrates to ubiquitinate. SMARCAL1 harbors an RPA2-binding motif to directly interact with RPA. ZRANB3 contains a PCNA-interacting protein (PIP) motif, an AlkB homolog 2 PCNA interacting motif (APIM) and a NPL4 zinc-finger (NZF) motif that cooperate to bind polyubiquitinated PCNA. The HIRAN, HARP and SRD (substrate recognition) domains of HLTF, SMARCAL1, and ZRANB3, respectively, enable structure specific DNA binding. The HNH motif of ZRANB3 is a nuclease domain.
