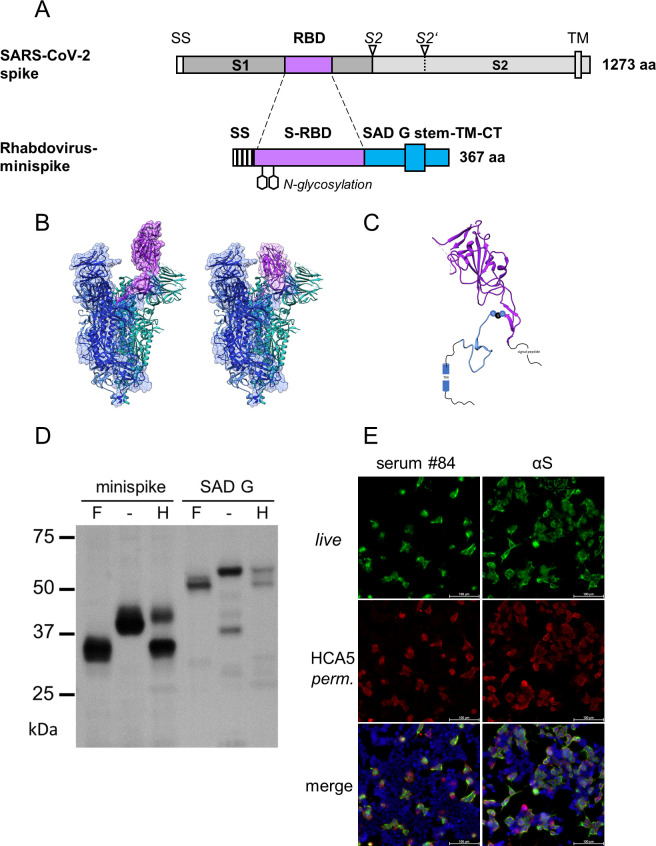
Fig 1. Design and expression of minispike.
(A) Schematic representation of the SARS-CoV-2 spike protein and of the chimeric minispike protein containing a hIgG signal sequence (SS) the SARS-CoV-2 RBD (purple), and the RABV G stem/anchor sequence (blue). Two consensus N-gylcosylation sites are indicated. S2 and S2′ arrowheads indicate protease cleavage sites, TM transmembrane domain. (B) Ribbon model of the SARS-CoV-2 S protein in the RBD “up” (PDB 6VYB) and “down” (PDB 6VXX) conformation with RBD residues included in the minispike protein highlighted in purple. The EM density map is shown in grey. (C) Model of the chimeric minispike construct. Elements with available structural information are shown as ribbon diagrams and include the RBD of SARS-CoV-2 (purple, PDB 6VXX) and parts of the RABV G-protein (blue, PDB 6LGX). The GSG Linker connecting the two domains is depicted as blue (G) and black (S) circles. Elements of unknown structure including signal peptide and C-terminus of RABV G are shown as black lines. A blue cylinder (TM) indicates the transmembrane domain. (D) Complex N glycosylation of minispike protein. Extracts from HEK293T cells transfected with pCR3-minispike and RABV G as a control were treated with PNGase F (+F), which cleaves off all N-linked oligosaccharides, left untreated (-) or treated with Endoglycosidase H (+H), unable to cleave complex sugars. The minispike protein acquires EndoH-resistant complex sugars, indicating transport through the Golgi apparatus. Proteins were visualized by incubation with HCA-5 serum, recognizing the common C-tail. (E) Surface expression and recognition of minispike by COVID-19 patient serum. Live, unpermeabilized HEK293T cells transfected with pCR3-minispike were first stained with a representative COVID-19 convalescent serum at 1:300 dilution (left panel) or conformation-specific SARS-CoV S Mab CR3022 (right panel), and anti-human IgG/AlexaFluor488 (green). Following fixation with 4% paraformaldehyde (PFA) and permeabilization with 0,1% Saponine, cells were in addition stained with HCA-5/anti-rabbit AlexaFluor555 (red) recognizing the intracellular RABV C-tail. Cell nuclei were visualized with ToPro3-iodide (blue). Size bar indicates 100 μM.