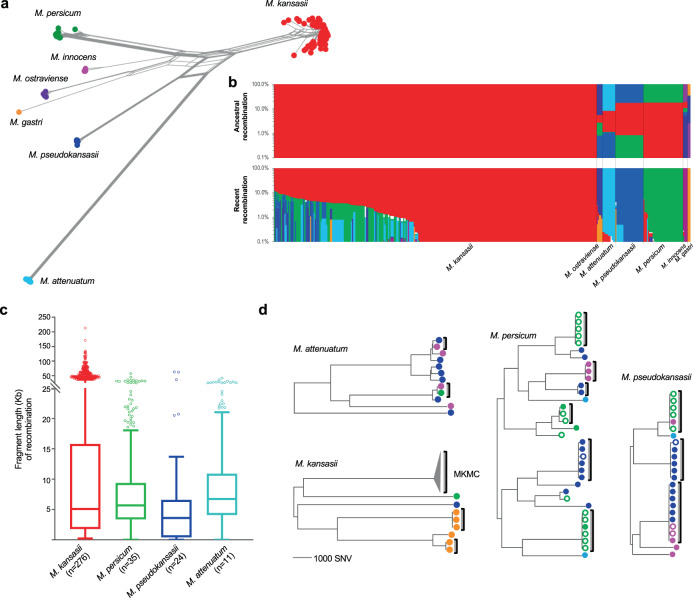
Fig. 2. Genomic recombination and its contribution in speciation and diversification of the M. kansasii complex.
a Phylogenetic network of the M. kansasii complex based on the core genome alignment of 358 isolates. b Population structure and genomic recombinations inferred by fastGEAR. Each line represents the genomic constitution (exhibited as color strips) of individual isolates according to ancestral (upper panel) or recent (lower panel) recombinations. Strip colors represent the different species as in panel (a). White strips (lower section) represent recent recombinations from unknown sources. Source data are provided as a Source Data file. c Length distribution of recombinant fragments in the four major species. Boxes show the median and interquartile range (IQR) while whiskers extend to a maximum of 1.5× IQR. d Maximum likelihood phylogeny of the four major species based on non-recombinant SNVs. Brackets indicate clusters containing isolates with an average pair-wise genomic difference of fewer than 100 SNVs. The colors of terminal nodes indicate the geographical origins of the isolates, corresponding to Fig. 1a. Filled circles indicate a human source; empty circles, an environmental source. MKMC M. kansasii main cluster.