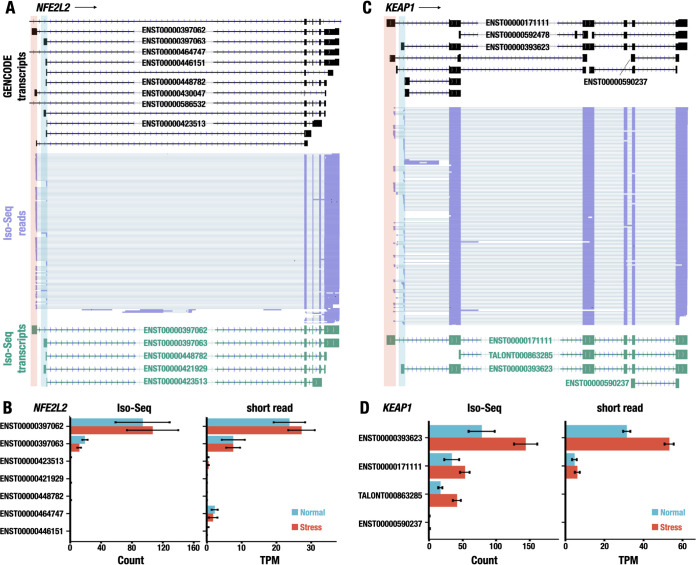
FIG 3.
Identification of alternative TSS usage in NFE2L2 and KEAP1 genes. (A and C) Representative Iso-Seq reads aligned to NFE2L2 (A) and KEAP1 (C) genes. The transcript structures in the GENCODE database are represented with black (GENCODE transcripts). The reads of Iso-Seq analysis are shown in purple (Iso-Seq reads). The transcript models made from Iso-Seq data are shown in green (Iso-Seq transcripts). The distal and proximal first exons are highlighted in red and blue, respectively. The ensemble IDs (with prefix of ENST) for transcripts annotated in the GENCODE database or the temporary IDs (with prefix of TALONT) for transcripts not annotated in the database are shown on the transcript models. (B and D) Expression levels of transcript isoforms of NFE2L2 (B) and KEAP1 (D) genes detected by Iso-Seq (left) and/or short-read-based RNA-Seq (right). The expression levels under normal and stress conditions are represented. The values are shown as the means ± SD.