Figure 1.
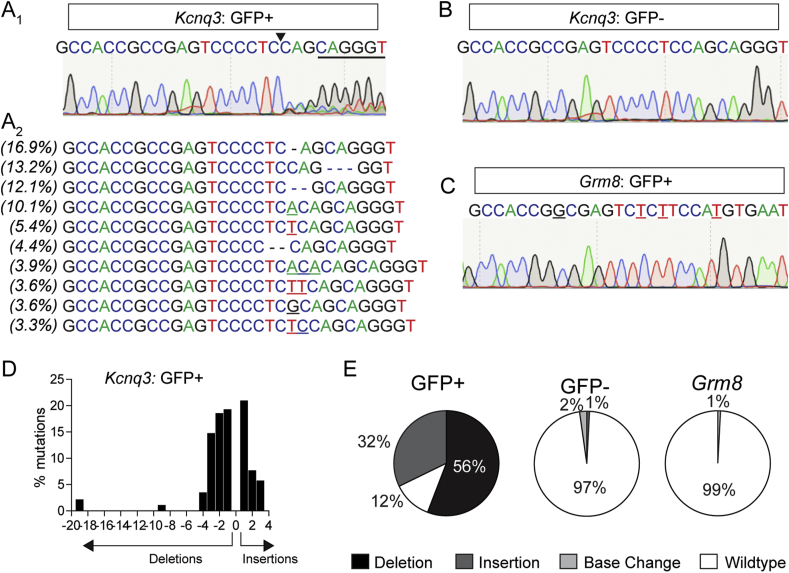
Analysis of targeted KCNQ3 mutagenesis in DATCremice. Analysis of FACS-sorted GFP+ (A) and GFP- (B) nuclei from mice co-injected with AAV1-FLEX-SaCas9-U6-sgKCNQ3 and AAV1-FLEX-EGFP-KASH into the VTA. (A1) sgKcnq3 sequence with PAM underlined and SaCas9 cut site indicated by black arrow. Chromatogram results display multiple peaks beginning at the cut site. (A2) Top 10 mutations at the cut site with the percent of total reads for which they occur on the left. Base changes: bolded. Insertions: underlined. Deletions: marked with a “-” (dash). (B) Chromatogram of GFP- nuclei sequencing displaying no evidence of mutations at the SaCas9 cut site. (C) Chromatogram of sgKcnq3 predicted off-target in Grm8 displaying no evidence of mutations at the SaCas9 cut site. (D) Frequency distribution of insertions and deletions in Kcnq3 from GFP+ nuclei. (E) Percent of wild-type, deletions, insertions, and base changes as percent of total reads for Kcnq3 in GFP+ (left) and GFP- nuclei (middle) and the predicted off-target Grm8 (right).