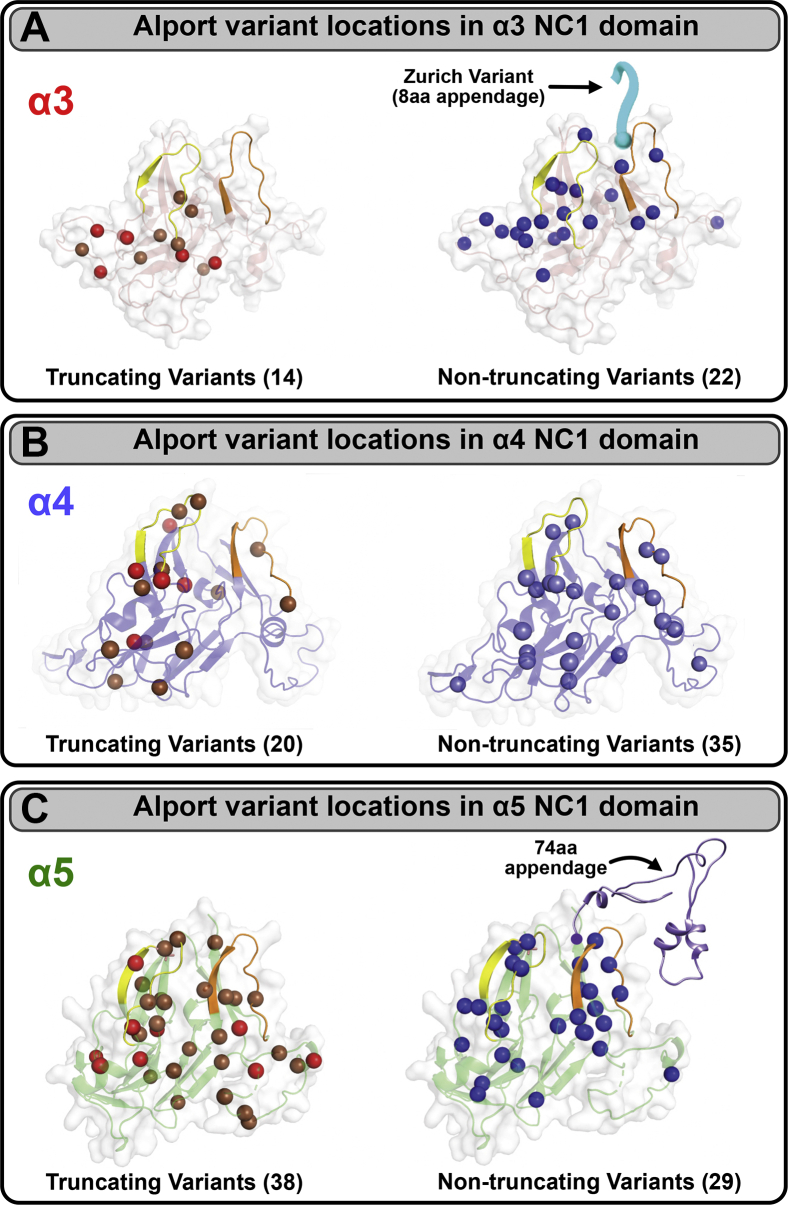
Figure 10.
Crystal structure of the α345 hexamer reveals the location of known Alport variants in the α3, α4, and α5 NC1 domains.A, distinct pathogenic Alport variants were mapped within the 3D structure of the α3 NC1 domain (source: HGMD 2020.1). The total number of truncating (small deletion, brown; nonsense, red) and nontruncating (missense variants, blue) variants are shown in parentheses. Truncating variants result in the premature stop codon and expression of the truncated form of COL4A3, which does not incorporate into the GBM (left). Although nontruncating missense variants (blue) do not affect the overall length of the NC1 domain (right), they may result in conformational changes of crucial regions within NC1. These nontruncating variants can incorporate into the GBM but are functionally defective. The Zurich variant (cyan) is a combination of a small deletion and insertion, resulting in an 8-aa appendage to the NC1 domain. Therefore, it belongs to “nontruncating” variant subgroup (right). B, distinct pathogenic Alport variants were mapped within the 3D structure of the α4 NC1 domain (source: HGMD 2020.1). Color codes are as in panel A. C, distinct pathogenic Alport variants were mapped within the 3D structure of the α5 NC1 domain (source: HGMD 2020.1). A distinct Alport variant, 74-amino acid appendage at the C-terminus of α5 chain, is depicted on the right. Color codes are as in panel A. For an expanded description of the genetic identity of the α3, α4, and α5 variants, see Figs. S12–S14. GBM, glomerular basement membrane.