Figure 1.
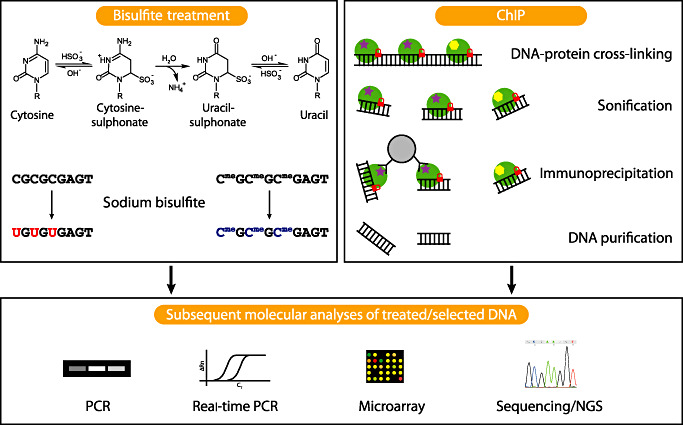
Commonly used methods to analyze epigenetic alterations. Sodium bisulfite treatment, which is frequently used for the investigation of DNA methylation, converts cytosine to uracil, while 5‐methylcytosine (5mC) and 5‐hydroxymethylcytosine are protected against this conversion. Subsequent analyses of the bisulfite‐treated DNA using PCR [methylation‐specific PCR (MSP)], real‐time PCR (MethyLight), microarrays, different sequencing approaches or mass spectrometry (MassARRAY EpiTyper assay, not depicted) allow to distinguish cytosines converted to uracil from unaltered cytosines. Chromatin immunoprecipation (ChIP) is frequently used to investigate specific histone modifications. Therefore, histones are stably cross‐linked to DNA by formaldehyde, in vivo. After cell lysis, DNA is fragmented by sonification. Antibodies against the modified histone of interest are added, bind to the modified histone and are afterwards pulled down by, for example, protein A/G coated agarose beads (protein A/G is capable of binding the Fc fragments of the specific antibodies). Thereafter, DNA protein cross‐links are reversed, and the DNA that was bound to the modified histone is purified for subsequent analyses. PCR and real‐time PCR focus on specific DNA regions, whereas microarray based methods (ChIP‐on‐chip) or next generation sequencing methods (ChIP‐seq) enable whole genome coverage. Histones are symbolized by green circles; two different histone modifications are symbolized by a purple star and a yellow hexagon, respectively; DNA‐protein cross‐linking is indicated by a red lock; the grey circle represents an agarose bead with protein A/G coating.
