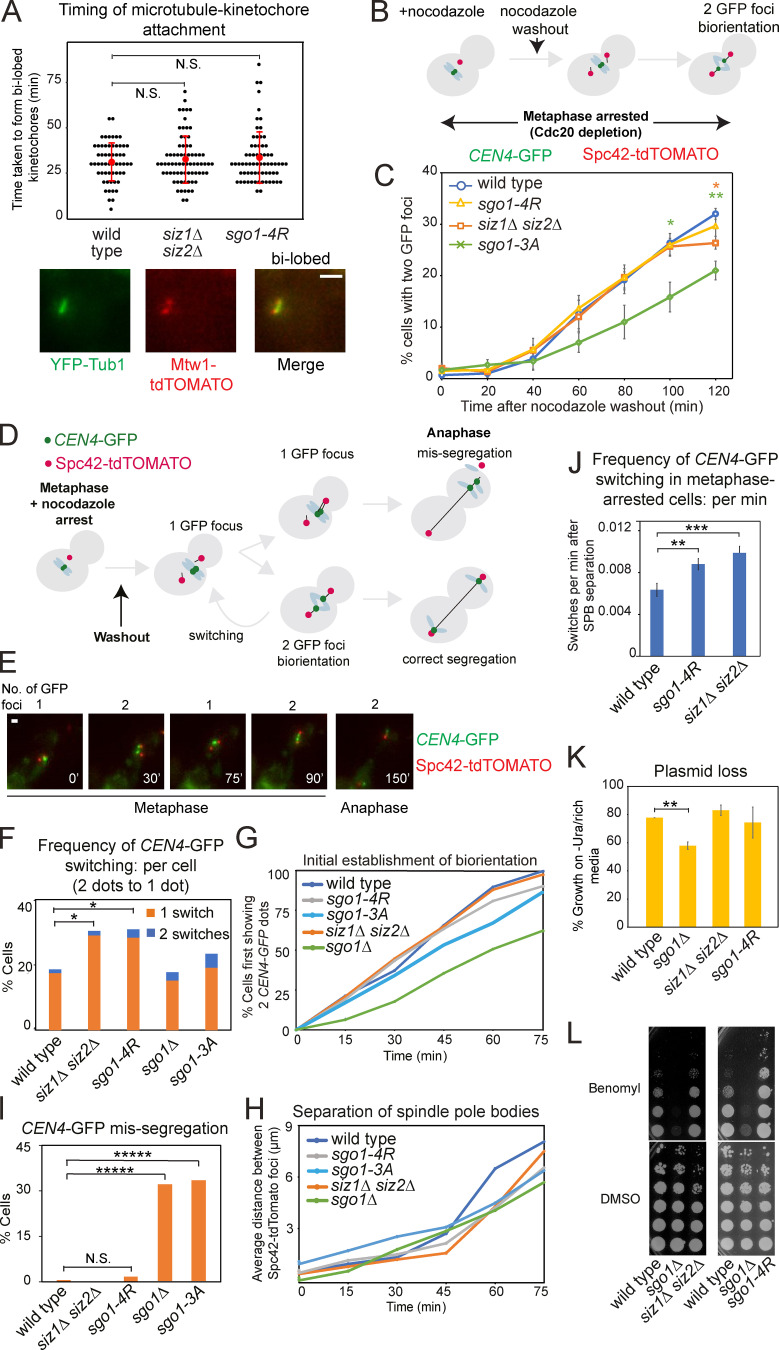
Figure 6.
SUMOylation of Sgo1 facilitates stable microtubule–kinetochore attachments. (A) Siz1/Siz2-mediated SUMOylation does not impair the initial microtubule–kinetochore attachment. Strains used carried pMET-CDC20 YFP-TUB1 and MTW1-tdTOMATO, and were wild type (AMy29568), siz1Δ siz2Δ (AMy29784), and sgo1-4R (AMy29782). Cells were arrested in metaphase by Cdc20 depletion and nocodazole treatment. After nocodazole washout, cells were imaged every 5 min while maintaining Cdc20 depletion. Images were acquired as described in Materials and methods and were maximum-intensity projected for Mtw1-tdTOMATO and YFP-Tub1. Scale bar = 1 µm. The timing of bilobed kinetochore formation (as shown in the representative image) was measured. At least 50 cells from two independent experiments were analyzed for each strain. Error bars represent standard deviation, with the red dots indicating mean values. Statistics: two-tailed Student’s t test. N.S., not significant. (B and C) SUMO-deficient Sgo1 is proficient for sister kinetochore biorientation. (B) Experimental scheme for evaluating sister kinetochore biorientation after nocodazole washout. Briefly, cells were arrested in metaphase by Cdc20 depletion in the presence of nocodazole. After drug washout, cells remained arrested in metaphase and were fixed and visualized as described in Materials and methods. (C) sgo1-4R and siz1Δ siz2Δ cells are proficient in the initial establishment of sister kinetochore biorientation. Strains used carried pMET-CDC20, CEN4-GFP, and SPC42-tdTOMATO and were wild type (AMy4643), sgo1-3A (AMy8923), siz1Δ siz2Δ (AMy27803), and sgo1-4R (AMy26278). Typically, 200 cells were analyzed for each strain in each experiment. Shown are the average values of three independent experiments, with error bars representing standard error. Statistics: two-tailed Student’s t test comparing to the wild-type strain (*, P < 0.05; **, P < 0.01). (D–I) Unstable kinetochore–microtubule attachments in SUMOylation mutants. Strains used in C, together with sgo1Δ (AMy6117), were monitored by live imaging on a microfluidics device. (D) Scheme describing biorientation assay after nocodazole washout. Briefly, nocodazole was washed out from metaphase-arrested cells, and state of CEN4-GFP was tracked by live-cell imaging. (E) Representative images of reassociation of two CEN4-GFP dots. Images were acquired as described in Materials and methods and were maximum-intensity projected for CEN4-GFP and Spc42-tdTOMATO. Scale bar = 1 µm. (F) Biorientation is unstable in siz1Δ siz2Δ and sgo1-4R cells. At least 50 cells from three independent experiments were analyzed for each strain. Shown are the percentages of cells with the indicated number of switchings from two CEN4-GFP dots back to one dot. Statistics: Fisher’s exact test (*, P < 0.05). (G) The initial establishment of biorientation is unaffected in siz1Δ siz2Δ and sgo1-4R cells. The time point at which a cell first displayed two CEN4-GFP dots was defined as the timing of the initial establishment of biorientation. (H) The distance between Spc42-tdTOMATO dots was measured by ImageJ, and the average distance was calculated for each time point. (I) siz1Δ siz2Δ and sgo1-4R cells do not show increased missegregation of chromosomes. The inheritance of CEN4-GFP by the daughter cells was assessed. Statistics: Fisher’s exact test (*****, P < 0.00001). (J) Switching frequency was increased in cells arrested in metaphase. Wild-type, sgo1-4R, and siz1Δ siz2Δ strains used in Fig. 6, D–I, were released from α-factor into methionine-containing medium (metaphase arrest). Cells were imaged every 15 min, and switching frequency was calculated as number of switches observed per minute cells spent after spindle pole body (SPB) separation. At least 100 cells from two independent experiments were evaluated for each strain. Error bars represent standard error. Statistics: two-tailed Student’s t test comparing to the wild-type strain (**, P < 0.01; ***, P < 0.001). (K) siz1Δ siz2Δ and sgo1-4R cells retain a CEN-containing plasmid to a similar extent as the wild-type cells. Wild type (AMy1176), sgo1Δ (AMy827), siz1Δ siz2Δ (AMy7625), and sgo1-4R (AMy21705) were transformed with a CEN-containing plasmid pRS316. Plasmid loss was evaluated in three independent transformants of each strain as described in Materials and methods. Error bars represent standard error. Statistics: one-tailed Student’s t test comparing to the wild-type strain (**, P < 0.01). (L) siz1Δ siz2Δ and sgo1-4R cells grew similarly as wild type or showed mildly improved growth on benomyl. Serially diluted cells of wild type (AMy1176), sgo1Δ (AMy827), siz1Δ siz2Δ (AMy7625), and sgo1-4R (AMy21705) were plated on medium containing 10 µg/ml benomyl or DMSO (solvent).