Table 2.
Starvation-regulated genes in T. brucei PCF
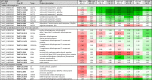 |
Selected transporters and enzymes upregulated in the absence of amino acids or glucose. Amino acid transporters of unknown and known (Tb927.8.8290, (33); Tb927.8.5450, (32); Tb927.8.4710, (31)) function, enzymes involved in proline catabolism, and enzymes of the TCA cycle not involved in proline metabolism. The table shows the fold change of mRNA levels between cells starved (2 h or 6 h) for amino acids (−AA) or glucose (−G) and nonstarved (+AA) PCF cells, for two biological replicates (I & II). The effect of FBS is shown by comparing cells grown in SDM79 + 10%FBS (FBS) with cells grown in starvation medium SDM79S including amino acids and glucose, but no FBS (+AA), i.e., FBS/+AA. Red shading indicates downregulation and green shading upregulation. For each 427_2018 gene identified we matched the corresponding syntenic 927 gene, if possible, and included the gene name and product description for convenience. For amino acid transporters we included the locus-based nomenclature (47) instead of the gene name.
