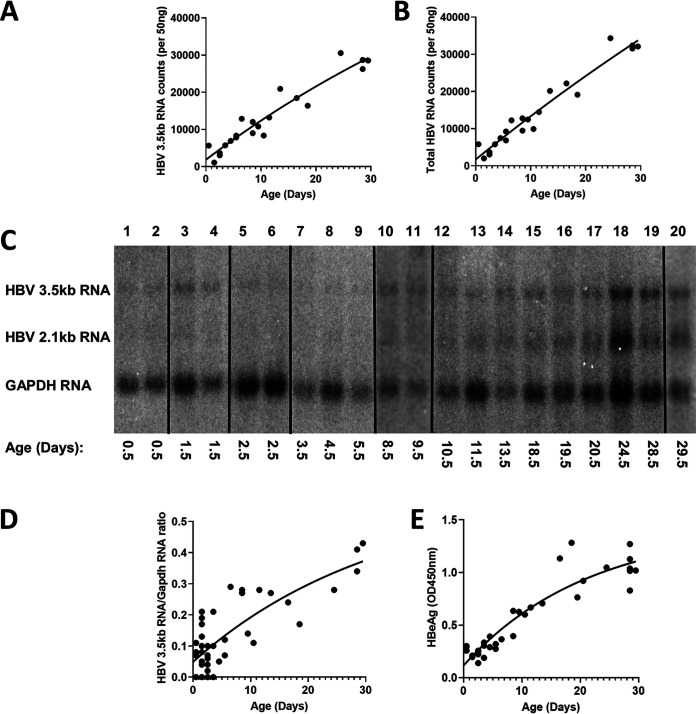
FIG 2.
Liver HBV transcripts and serum HBeAg levels throughout postnatal developmental of HBV transgenic mice. (A and B) Quantitative analysis of liver viral 3.5-kb RNA and total HBV RNA (3.5-kb plus 2.1-kb RNA), respectively, by NanoString gene expression analysis in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in gene expression throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4). (C) RNA (Northern) filter hybridization analyses of liver transcripts isolated from HBV transgenic mice of various postnatal developmental ages are shown. Noncontiguous lanes from a single analysis are presented. The probes used were HBVayw genomic DNA plus glyceraldehyde-3-phosphate dehydrogenase (Gapdh) cDNA. The Gapdh transcript was used as an internal control for the quantitation of the HBV 3.5-kb RNA. (D and E) Quantitative analysis of RNA (Northern) filter hybridization of the HBV 3.5-kb transcript (D) and serum HBeAg (E) in the HBV transgenic mice. One-phase decay analysis was used to estimate the changes in HBV RNA gene expression and serum HBeAg throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4).