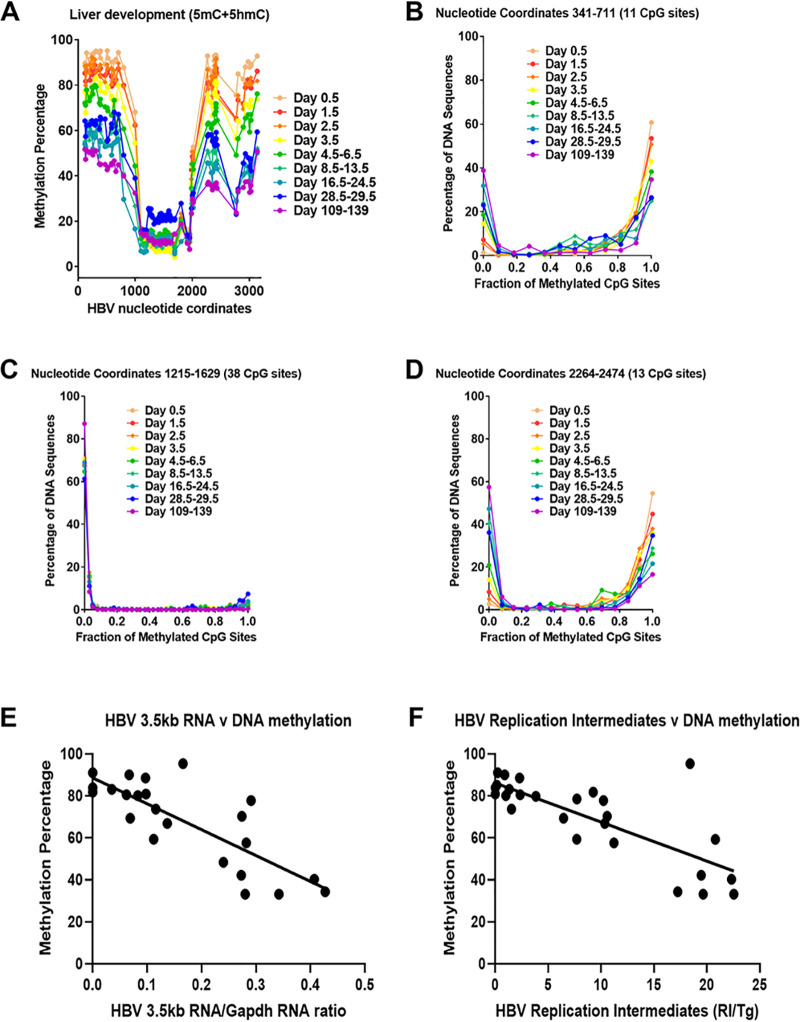
FIG 4.
Effect of postnatal liver development on HBV DNA methylation in HBV transgenic mice. (A) Percentage of CpG DNA methylation (5mC plus 5hmC; bisulfite treatment) at each position within the HBV genome from the livers of HBV transgenic mice throughout postnatal development is shown. The numbers of mice per group are 10 on day 0.5, 11 on day 1.5, 8 on day 2.5, 6 on day 3.5, 5 on days 4.5 to 6.5, 6 on days 8.5 to 13.5, 5 on days 16.5 to 24.5, 5 on days 28.5 to 29.5, and 2 on days 109 to 139. The viral transcription initiation sites for the X gene, the precore/pregenomic, the large surface antigen, and the middle/major surface antigen transcripts plus the CpG island are located at nucleotide coordinates 1310, 1785/1821, 2809, and 3159/3178 plus 1000 to 2000, respectively. (B to D) Effect of postnatal liver development on HBV DNA methylation distribution. The CpG DNA methylation (5mC plus 5hmC; bisulfite treatment) frequency distributions across the 11, 38, and 13 sites within HBV nucleotide coordinates 341 to 711, 1215 to 1629, and 2264 to 2474, respectively, for the viral genome from the livers of HBV transgenic mice throughout postnatal development are shown. The numbers of mice per group are 10 on day 0.5, 11 on day 1.5, 8 on day 2.5, 6 on day 3.5, 5 on days 4.5 to 6.5, 6 on days 8.5 to 13.5, 5 on days 16.5 to 24.5, 5 on days 28.5 to 29.5, and 2 on days 109 to 139. (E) Linear regression analysis was used to correlate HBV 3.5-kb RNA levels with the average viral DNA methylation percentage across HBV nucleotide coordinates 341 to 711 throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4; R2, 0.64; P < 0.001; y-intercept, 89%). (F) Linear regression analysis was used to correlate HBV DNA replication intermediate levels with the average viral DNA methylation percentage across HBV nucleotide coordinates 341 to 711 throughout postnatal liver development in the HBV transgenic mice (GraphPad Prism 8.4; R2, 0.59; P < 0.001; y-intercept, 86%).