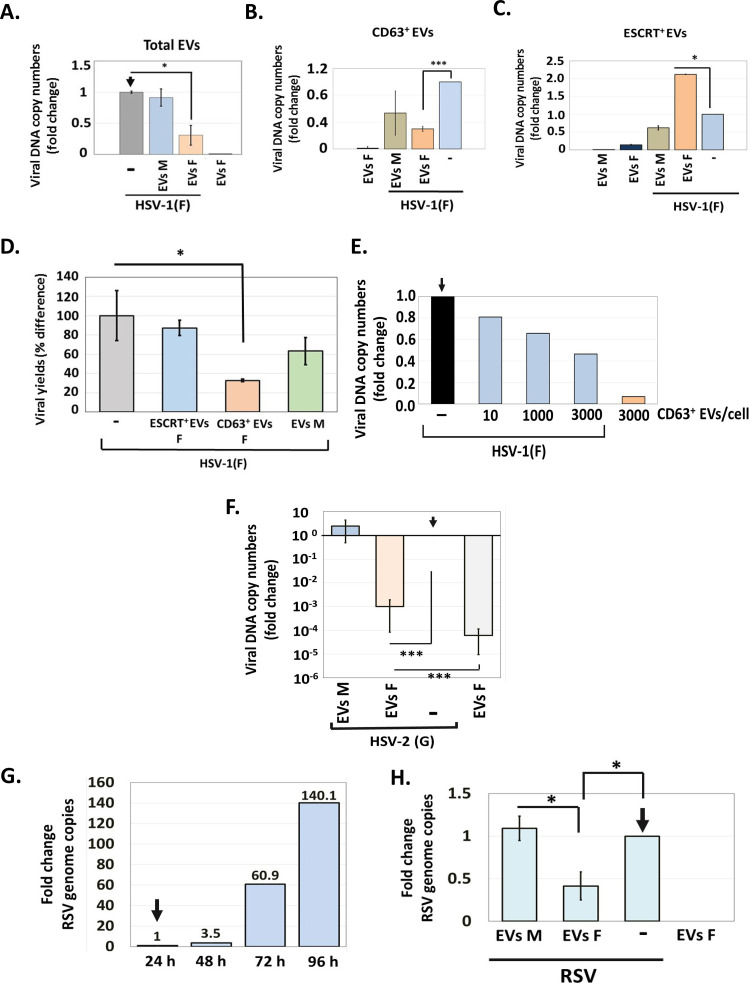
FIG 6.
Effect of EVs on HSV-1 infection. (A) EVs from infected or uninfected HEL cells present in the first seven fractions of the gradient described in the legend for Fig. 1 were quantified by NTA and used to expose uninfected HEL cells (1,000 EVs/cell). At 2 h post-EV exposure, the cells were infected with HSV-1(F) (0.01 PFU/cell). The cells were harvested at 24 h postinfection, and quantification of the viral genome was done by qPCR analysis as detailed in Materials and Methods. Cells exposed only to EVs or to virus served as controls. (B and C) CD63+ EVs (B) and ESCRT+ EVs (C) were isolated from infected and uninfected HEL cells at 24 h postinfection (0.01 PFU/cell), as for Fig. 1. Equal volumes of EVs were used to expose HEL cells for 2 h, followed by HSV-1 infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above. Relative fold gene expression was calculated using the delta-delta threshold cycle (CT) method. (D) CD63+ and ESCRT+ EVs were isolated from infected and uninfected HEL cells at 24 h postinfection (0.01 PFU/cell), as for Fig. 2. Equal volumes of EVs were used to expose HEL cells for 2 h, followed by HSV-1 infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and quantification of the infectious virus was done by plaque assay in Vero cells. (E) CD63+ EVs were isolated from HSV-1(F)-infected HEL cells as described above, and different doses were used to expose HEL cells for 2 h prior to HSV-1(F) infection (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above. Exposure to CD63+ EVs from infected cells served as a contamination control. Fold change is relative to infected but non-EV-treated cells. (F) HEL cells were exposed to CD63+ EVs isolated from HSV-1(F)-infected or uninfected cells as for panel B, but infection was done with HSV-2(G) (0.01 PFU/cell). The cells were harvested at 48 h postinfection, and viral genome copy numbers were quantified as described above using primer pairs targeting the gG region of HSV-2 genome. (G) HEL cells were infected with RSV (0.01 PFU/cell). The cells were harvested every 24 h up to 96 h postinfection. Total RNA was extracted, reversed transcribed, and used to quantify the RSV genome. (H) HEL cells were infected with RSV (0.01 PFU/cell) for 48 h, followed by exposure to CD63+ EVs derived from HSV-1(F)-infected or uninfected cells as for panel B for another 48 h. The cells were harvested at 96 h postinfection, and viral genome copy numbers were quantified as detailed in Materials and Methods. All values are derived from triplicate samples. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.