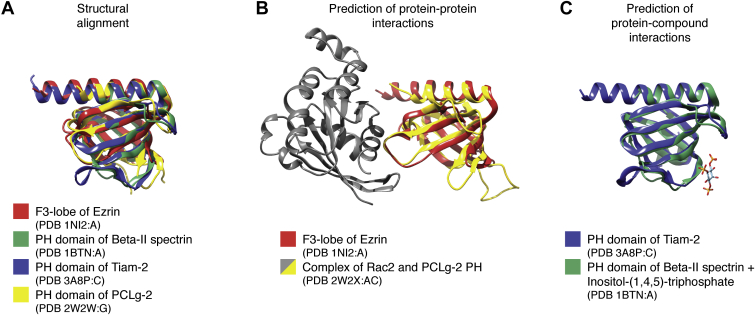
Figure 1.
Detecting protein–protein and protein–compound interactions with Structural Blast.A, F3 lobe of Ezrin from Protein Data Bank (PDB) 1ni2:A (red); pleckstrin homology (PH) domain of Beta-II spectrin from PDB 1btn:A (green); PH domain of Tiam-2 from PDB 3a8p:C (blue); and PH domain of PLCg-2 from PDB 2w2w:G (yellow). B, structure alignment of F3 lobe of Ezrin (1ni2:A, red) to the PDB complex (2w2x:AC) of Rac2 (gray) and PLCg-2 PH (yellow). PrePPI (35) uses this template complex to predict an interaction between Rac2 and Ezrin. C, the first PH domain of Tiam-2 is predicted by LT-Scanner (94) to bind inositol-(1,4,5)-triphosphate based on structure superposition of a homology model for Tiam-2 (blue) onto the PH domain of Beta-II spectrin (green) complexed with inositol-(1,4,5)-triphosphate (stick representation) (PDB: 1btn). Even though the sequence identity between the proteins is undetectable by pairwise global sequence alignment, the five residues specifically involved in binding inositol-(1,4,5)-triphosphate (102) are largely conserved in Tiam-2 as revealed by structure-based sequence alignment (K–K, R–R, S–R, Y–H, K–K, W–W).