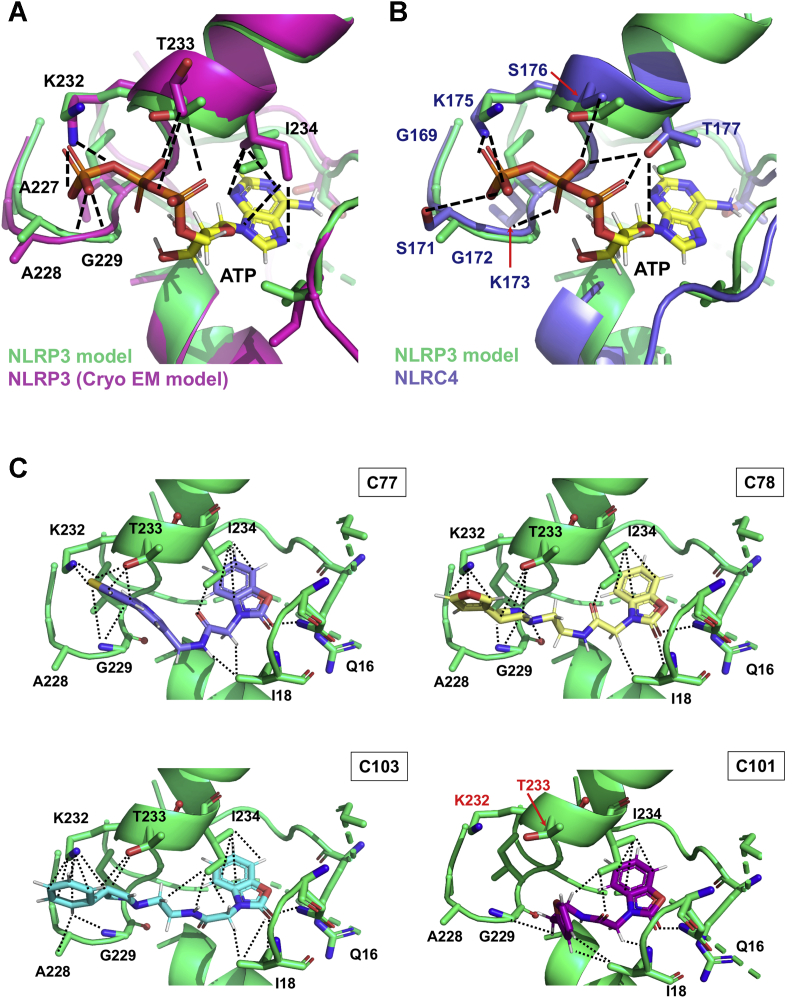
Figure 7.
Structural insights for the allocation of the adenine nucleotide ligands or the inhibitors within the ATP-binding site of the NACHT domain.A, structural alignment for the ATP-binding pockets of the NLRP3 model used for the virtual screening (green) and of the cryo-EM structure (magenta). B, structural alignment for the ATP-binding pockets of the NLRP3 model used for the virtual screening (green) and of the NLRC4 crystal structure. The ATP ligand is displayed in the CPK code with carbons in yellow. The lateral chains of the residues from the Walker A motif predicted to be involved in the ATP stabilization are highlighted in sticks and colored in CPK colors. Stabilizing interactions between residues from the NLRP3 cryo-EM structure (A) or NLRC4 (B) and ATP are shown as the dashed line. C, structural models for the allocation of C77 (light purple), C78 (pale yellow), C103 (cyan), and C101 (pink) within the ATP pocket of our NLRP3 model. Residues stabilizing the inhibitors are shown as sticks and colored in the CPK code. Interactions between side chains of these residues and the compounds are displayed as dotted lines. NACHT, nucleotide-binding and oligomerization; NLRC4, NLR family CARD domain–containing 4; NLRP3, nucleotide-binding oligomerization domain, leucine-rich repeat and pyrin domain–containing protein 3.