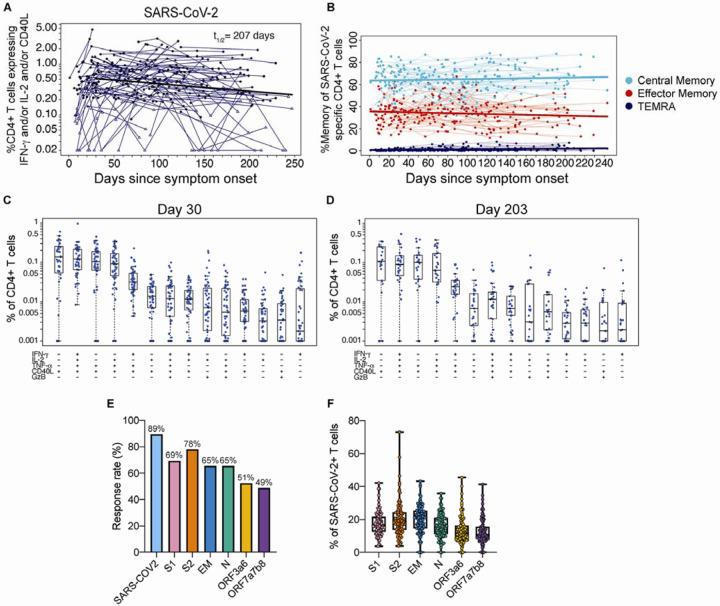
Figure 5. CD4+ T cell responses to SARS-CoV-2 antigens.
(A) The sum of background-subtracted CD4+ T cells expressing ex vivo IFN-γ, IL-2 and/or CD40L to peptide pools spanning SARS-CoV-2 structural proteins: S1, S2, envelope (E), membrane (M), nucleocapsid (N), and the following ORFs: 3a, 3b, 6, 7a, 7b, and 8 (n=114; tested in singlets) for each individual/timepoint. Each sample that is ‘positive’ (by MIMOSA) for at least one SARS-CoV-2 antigen is indicated by a solid circle, whereas samples that are ‘negative’ for all of the SARS-CoV-2 antigens at that timepoint are indicated by open triangles. The bold line represents the median fitted curve from a nonlinear mixed effects model of post-day 30 responses among those with a positive response at >= 1 time point; t1/2 is the median half-life estimated from the median slope, with 95%CI [104, 411]. (B) The proportion of SARS-CoV-2-specific CD4+ T cells expressing a specific memory phenotype over time: central memory (CCR7+ CD45RA−), effector memory (CCR7− CD45RA−) or TEMRA (CCR7− CD45RA+); restricted to ‘positive’ responders. Polyfunctionality of SARS-CoV-2-specific CD4+ T cells are shown at (C) 21–60 days since symptom onset (median, 30 days) and (D) >180 days median post symptom onset (median, 203 days). Percentages of cytokine-expressing CD4+ T cells are background subtracted and only subsets with detectable T cells are displayed. Data shown were restricted to ‘positive’ responders and a single data point per individual per time frame. All subsets were also evaluated for expression of IL-4, IL-5, IL-13, IL-17 and perforin, and were found to be negative. (E) Bar graphs indicate the proportion of COVID-19 convalescent patients who had a positive CD4+ T cell response to the individual SARS-CoV-2. peptide pool ex vivo stimulations. Some antigens were combined for stimulation as indicated. (F) For each subject with positive SARS-CoV-2- specific CD4+ T cells, the proportion of the total SARS-CoV-2 responding CD4+ T cells that are specific for each stimulation.