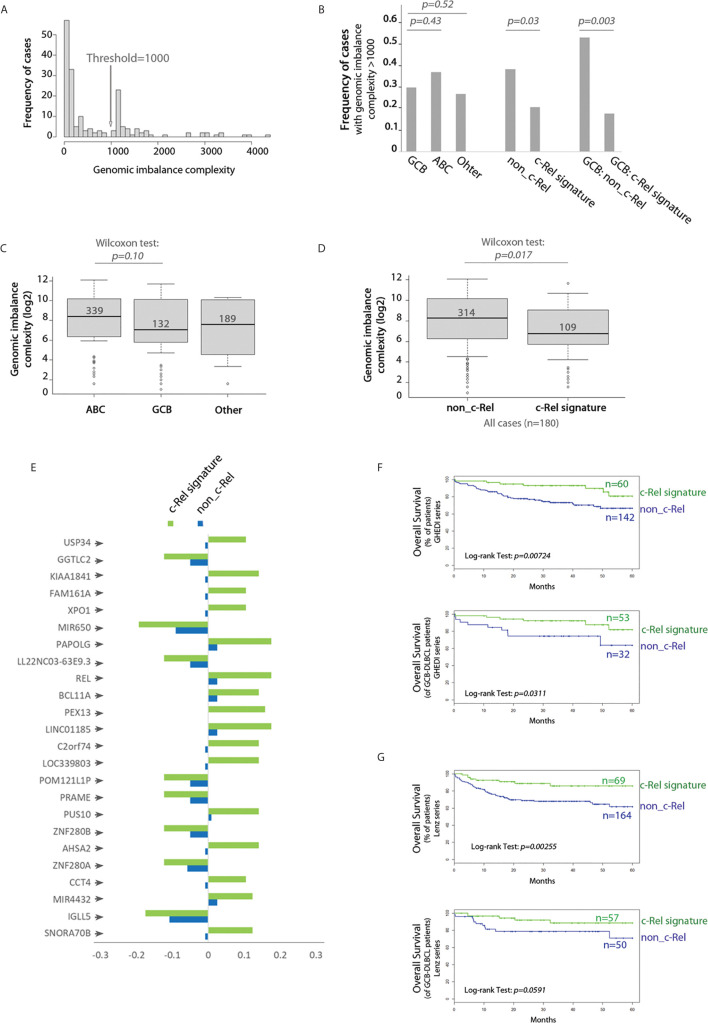
Figure 6.
c-Rel related gene copy number aberrations and overall survival. (A) Frequency histogram of the genomic imbalance burden reveals a bimodal distribution with a threshold at 1,000. (B) Frequency of DBCL cases with a genomic imbalance burden >1,000 according to the COO classification (left), absence (non_c-Rel) or presence (c-Rel signature) of the c-Rel signature (middle and right) among the entire GHEDI series (left and middle) or GCB-DLBCLs only (right). Chi2 test p-values are indicated on the top. (C) Boxplot of the genomic imbalance burden according to the COO classification. (D) Boxplot of the genomic imbalance burden according to the absence (non-c-Rel) or presence (c-Rel signature) of the c-Rel signature. (C, D) Median number of gene imbalances is indicated by the black solid line. Wilcoxon test p-values are shown on the top. (E) Frequency of cases with gene copy number aberrations for the 24 gene loci specifically imbalanced in DLBCL cases with the c-Rel signature. Negative values mean deletion while positive values mean amplification. (F, G) Kaplan–Meier overall survival curves of all (upper panel) or GCB (lower panel) patients from GHEDI and Lenz series respectively according to expression of the c-Rel signature (c-Rel signature, green curve) or not (non-c-Rel, blue curve). Number of cases is indicated on the right. The log-rank p-value is given within each graph.