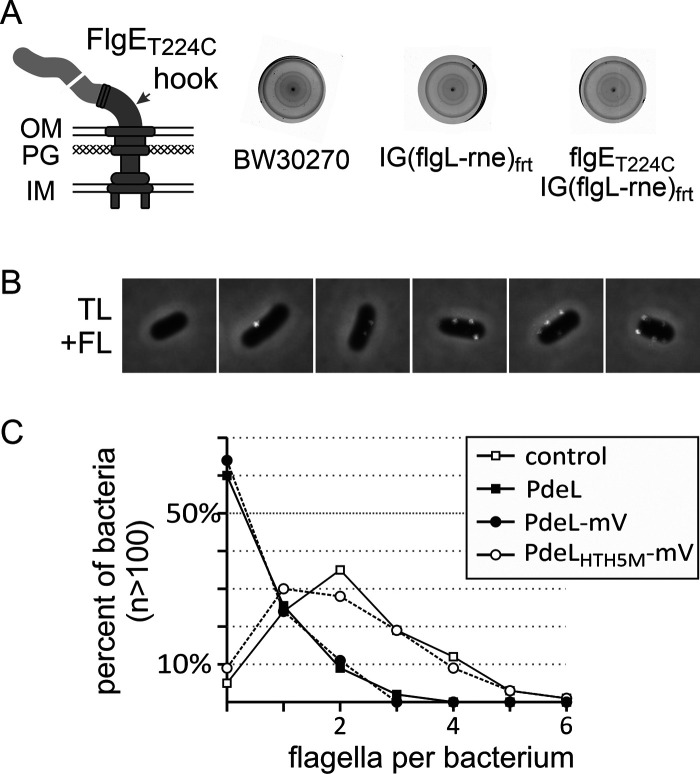
FIG 7.
Inhibition of flagellum synthesis by PdeL. The number of flagella per bacterium was determined by coupling the thiol-reactive fluorescent dye Alexa Fluor 568 C5 maleimide to the flagellar hook protein FlgET224C. (A) Motility of wild-type E. coli K-12 strain BW30270, strain T2611 carrying an FRT site in the intergenic region between flgL and the rne allele [IG(flgL-rne)FRT] (strain T2611), and its isogenic flgET224C mutant (strain T2614). Tryptone soft-agar plates were incubated for 4 h at 37°C. OM, outer membrane; IM, inner membrane; PG, peptidoglycan. (B) Representative microscopy images (overlays of transmitted light/phase contrast and red fluorescence [TL+FL]) of transformants of the flgET224C mutant strain T2614 stained with Alexa Fluor 568 C5 maleimide showing 0 to 5 fluorescent spots (in white). Each image represents 5 μm2. (C) Quantitative analysis of the number of fluorescent spots (flagella per bacterium) of transformants of flgET224C mutant strain T2614 with pKESK22 (control), Ptac pdeL provided by pKESL162 (PdeL), Ptac pdeL-mVenus (pKESL166) (PdeL-mV), and Ptac pdeLHTH5M-mVenus (pKESL209) (PdeLHTH5M-mV). Cultures of transformants were used to inoculate 15 ml LB medium with kanamycin to an OD600 of 0.08, and the cultures were grown at 37°C for 3 h and then harvested for flagellum staining and microscopy. Shown are the averaged data from three biological replicates, each based on the evaluation of fluorescent spots of >100 bacteria using ImageJ with the MicrobeJ plug-in.