Figure 3: Case study: a spatiotemporal multi-omics atlas of the aortic valve.
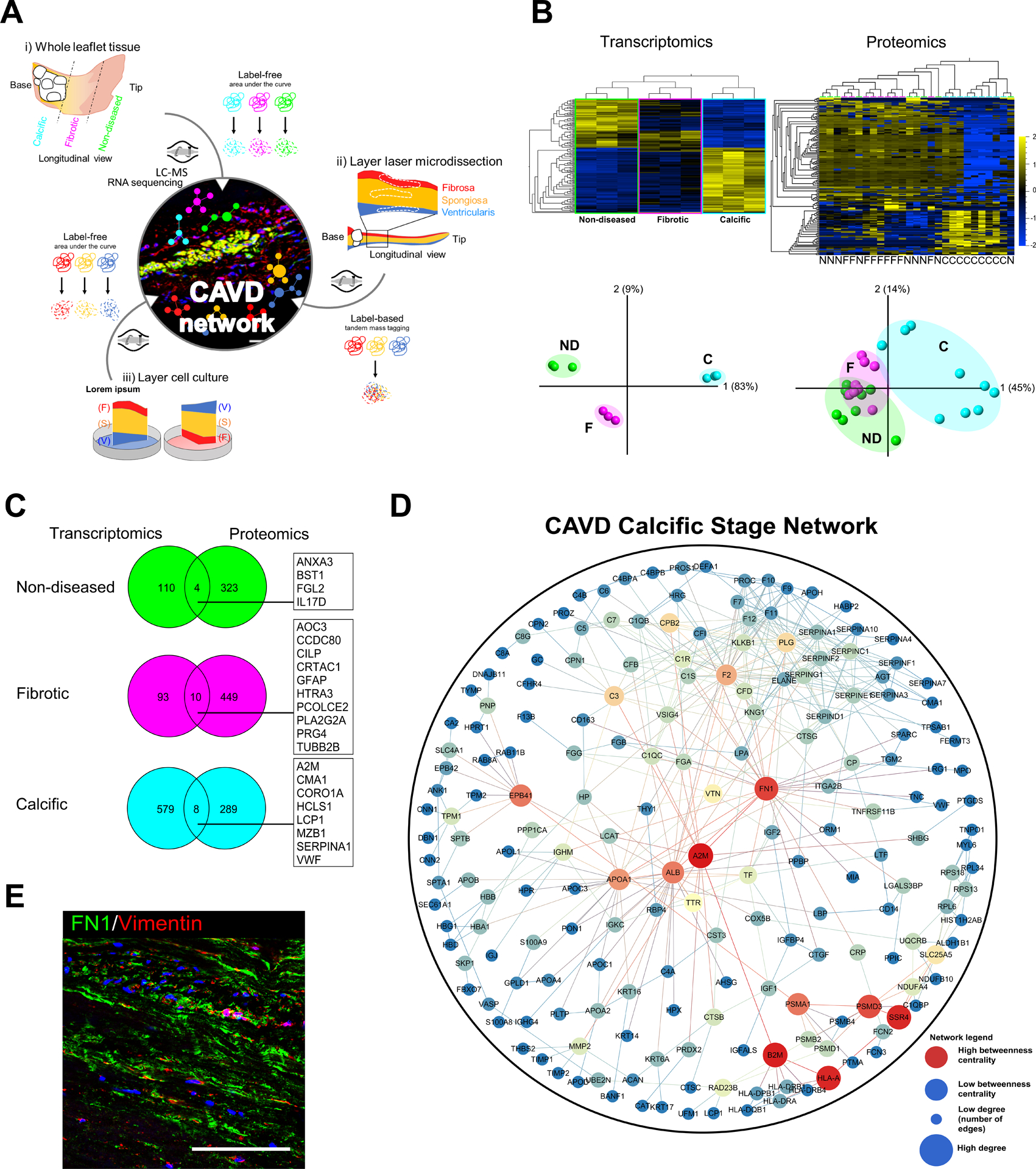
(A) Schlotter et al. employed proteomics and transcriptomics studies to investigate layer-specific pathobiology and temporal pathogenesis of human calcific aortic valve disease (CAVD). (B) Transcriptomics and proteomics leveraged to assess differences in molecular phenotypes across stages of disease (ND: non-diseased, F: fibrotic, C: calcified) demonstrate pathobiological differential enrichment and stage-specific clustering by principle component analyses (PCA). (C) Multi-omics integration in the aortic valve must be approached with care and sophistication, as the low cell density (and resultant abundance of proteinaceous extracellular matrix) and other regulatory or pathological mechanisms can lead to low correlation between tissue-based transcript and protein levels. (D) Network medicine approaches can be subsequently employed for target prioritization: protein-protein interaction (PPI) networks build from stage-specific proteomics of human aortic valves revealed key “hub node” proteins (red) implicated in valvular calcification. (E) Immunofluorescence staining in calcified regions of human aortic valves to validate the network-derived target FN1 (in green, nuclei in blue, VIC marker vimentin in red); scale bar = 100 μm. Adapted from26.
