Figure 1.
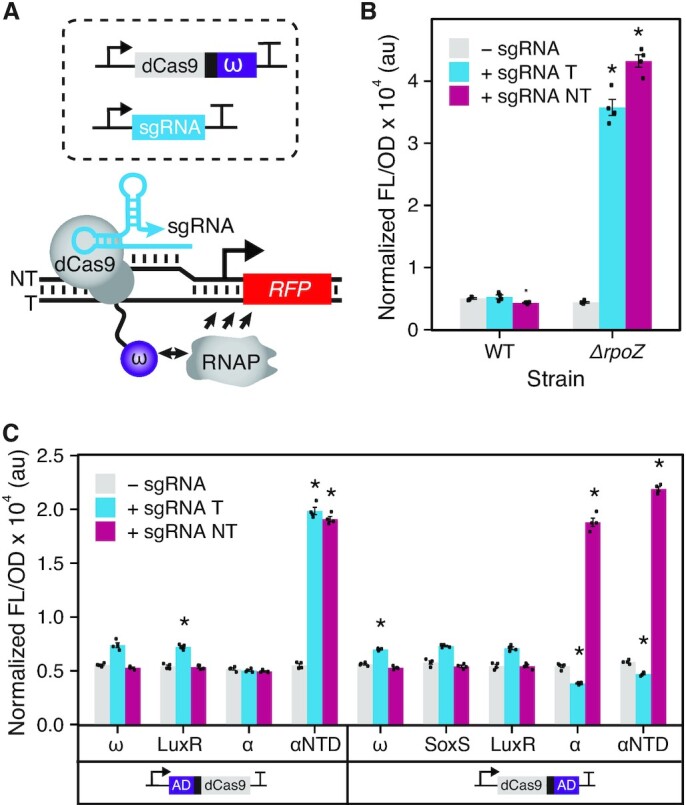
Synthetic transcription activation with dCas9 fusions to ADs. (A) Schematic of a CRISPRa system composed of dCas9 fused to the ω subunit of RNAP (dCas9-ω). For characterization, dCas9-ω in complex with the sgRNA was targeted upstream of a promoter driving RFP expression. Activation is achieved by local recruitment of RNAP to promoter elements. (B) Fluorescence characterization of a dCas9-ω system in wild-type strain E. coli K-12 BW25113 (WT) and a modified strain that lacks the gene encoding the ω subunit (ΔrpoZ). (C) Fluorescence characterization of dCas9 with N- or C-terminal fusions to different ADs through a two-alanine linker in E. coli MG1655 strain. ADs are derived from RNAP subunits: ω, α and αNTD; or transcription factors: SoxS and LuxR. Fluorescence measurements (measured in units of fluorescence [FL]/optical density [OD] at 600 nm) were performed with E. coli cells transformed with an RFP reporter plasmid, a plasmid encoding the dCas9 fusions, and a sgRNA-encoding plasmid or a no-sgRNA control plasmid. sgRNA variants used targeted PAMs located at 80 bp upstream of the promoter TSS on the template strand (+ sgRNA T) or 81 bp upstream on the non-template stand (+ sgRNA NT). Data represent mean values and error bars represent s.d. of n = 4 biological replicates. A two tailed Student's t test was used to calculate P value comparing against the no-sgRNA control. * P < 0.0001; P > 0.0001 has no asterisk.
