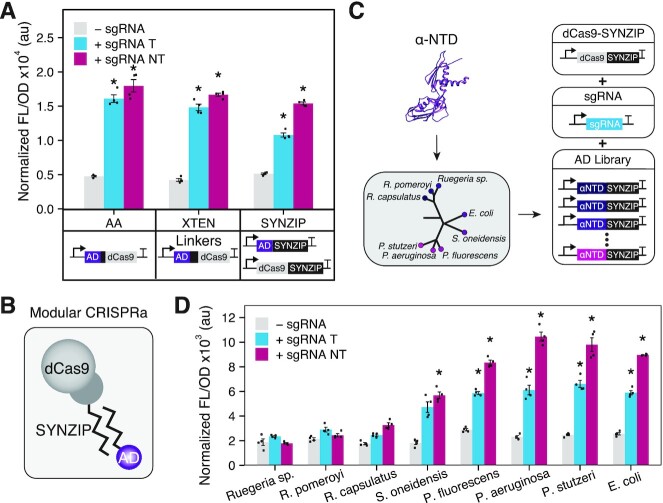
Figure 2.
Evaluation of different ADs and fusion strategies. (A) Characterization of different protein fusion strategies to recruit the αNTD AD through dCas9 by a covalent two-alanine (AA) linker, covalent 16 amino acid XTEN linker, or a noncovalent SYNZIP interaction domain. Schematics of DNA constructs shown under each data set. (B) Schematic of the modular CRISPRa design. The dCas9 and AD are independently fused to SYNZIP domains that form heterodimers. (C) A schematic illustrating creation and screening of an AD library using our modular CRISPRa platform. Different ADs were identified from αNTDs derived from 8 different bacterial species and translationally fused to a SYNZIP domain. These AD-encoding plasmids were then co-transformed with dCas9-SYNZIP and sgRNA-encoding plasmids to create new CRISPRa systems. (D) Characterization of the AD library. Fluorescence characterization (measured in units of fluorescence [FL]/optical density [OD] at 600 nm) was performed with MG1655 E. coli cells transformed with an RFP reporter plasmid, a dCas9-AD plasmid or separate dCas9- and AD-encoding plasmids, and a sgRNA-encoding plasmid or a no-sgRNA control plasmid. sgRNA variants used targeted PAMs located at 80 bp upstream of the promoter TSS on the template strand (+ sgRNA T) and 81 bp upstream on the non-template stand (+ sgRNA NT). Data represent mean values and error bars represent s.d. of n = 4 biological replicates. A two tailed Student's t test was used to calculate p value comparing against the no-sgRNA control. * P < 0.0001; P > 0.0001 has no asterisk.