Figure 3.
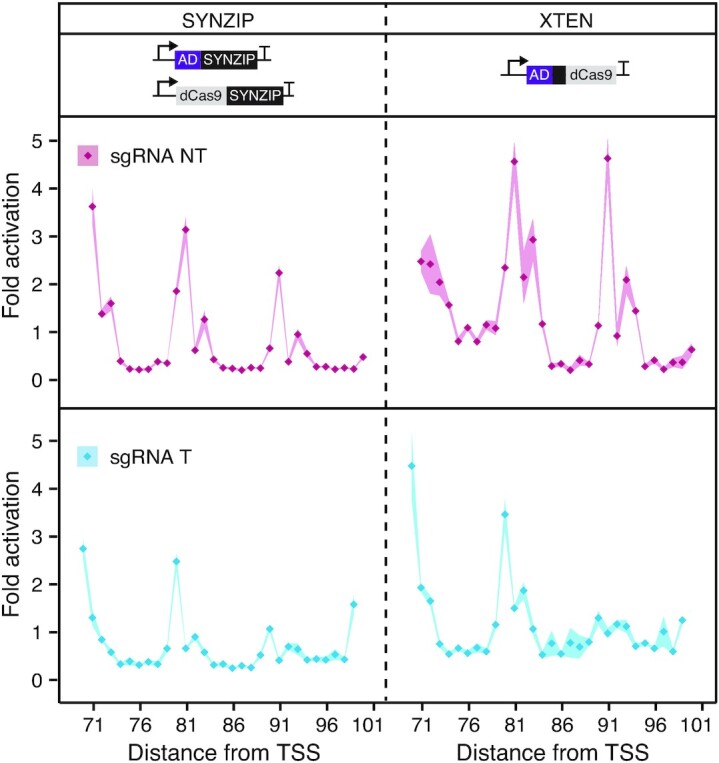
Characterization of distance-dependent activation patterns. Measuring the distance-dependent activation patterns of CRISPRa systems using XTEN and SYNZIP linkers. These systems were targeted to PAMs located between 71 and 101 bp upstream of the promoter TSS on both the template and non-template strands. Fluorescence characterization (measured in units of fluorescence [FL]/optical density [OD] at 600 nm) was performed in MG1655 E. coli cells transformed with an RFP reporter plasmid, a dCas9 plasmid, an AD plasmid and a sgRNA-encoding plasmid or a no-sgRNA control plasmid. Fold activation was calculated by dividing the [FL]/[OD] obtained in the presence of a targeting sgRNA against the no-sgRNA control within each reporter plasmid. Data represent mean values and shading represent s.d. of n = 4 biological replicates.
