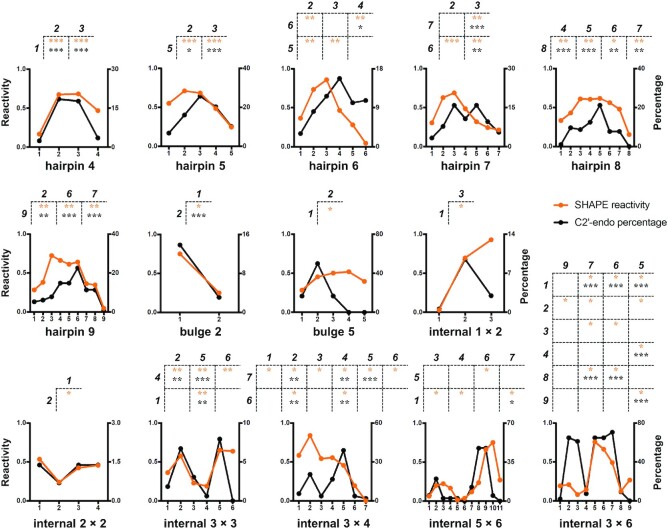
Figure 2.
Characteristic SHAPE patterns for hairpin, internal, and bulge loop motifs. The averaged SHAPE reactivities at all positions of each loop motif in benchmark RNAs are shown by orange lines. The proportions of the C2′-endo sugar pucker conformation (for RNAs in the non-redundant PDB list with 4.0 Å cutoff) are shown by black lines. The characteristic SHAPE patterns of each loop motif are shown at the top of each subplot, with the residues at the top of each table showing higher SHAPE reactivities than the residues to the left, and their P-values are indicated by asterisks (*0.01 < P-value ⩽ 0.05; **0.001 < P-value ⩽ 0.01; ***P-value ⩽ 0.001). The residue position is counted from the 5′ end of each loop motif. Detailed P-values and sample sizes are listed in Supplementary Tables S3 and S4.