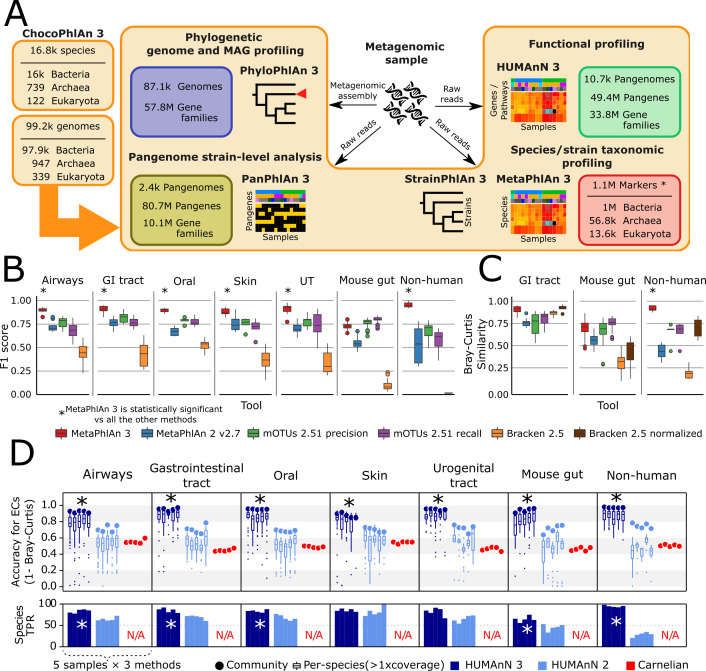
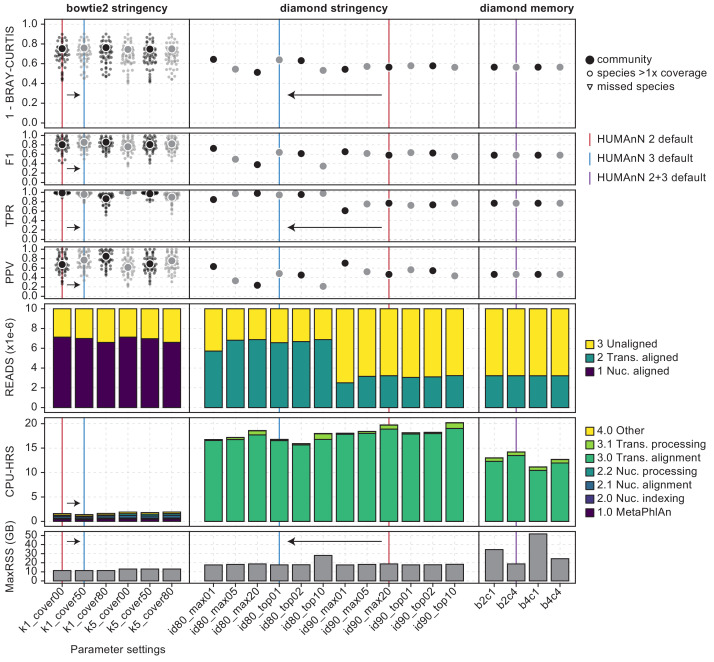
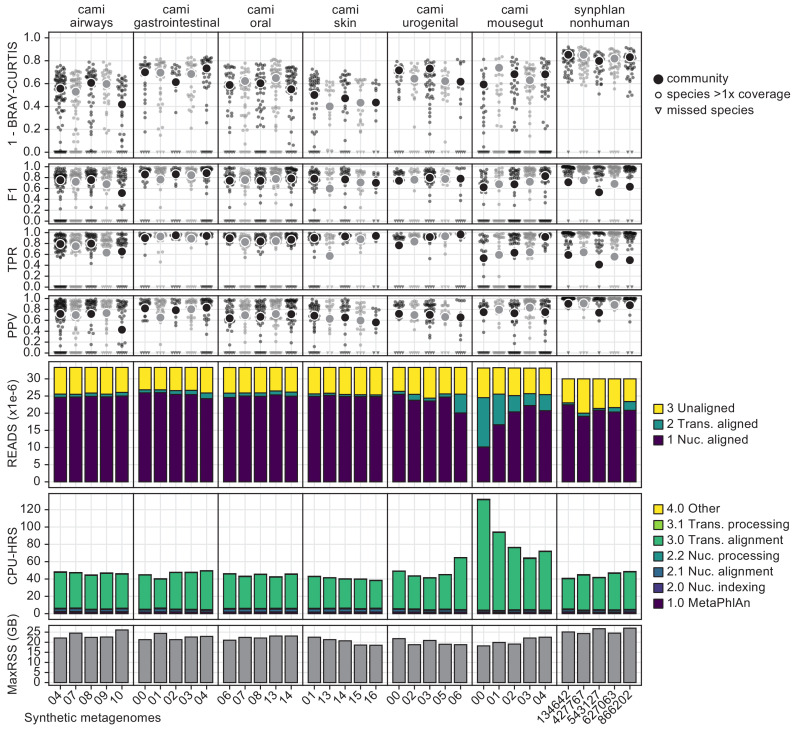
Figure 1. bioBakery 3 includes new microbial community profiling approaches that outperform previous versions and current methods.
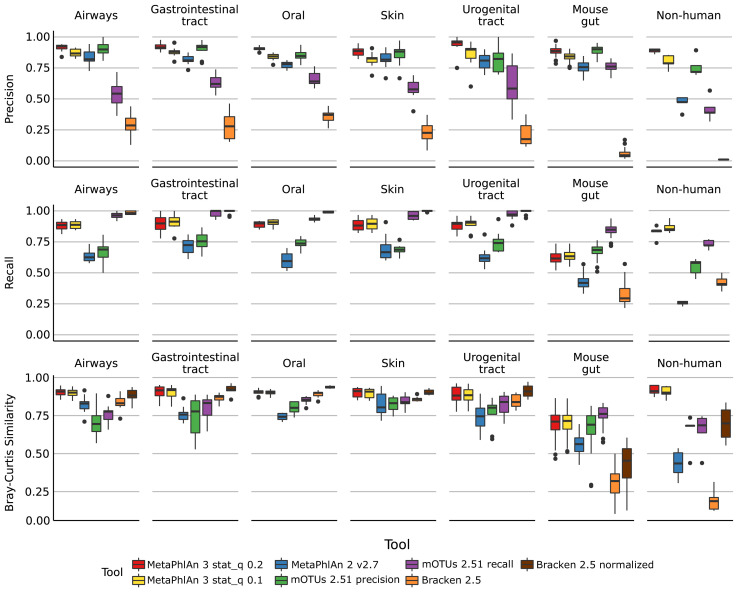
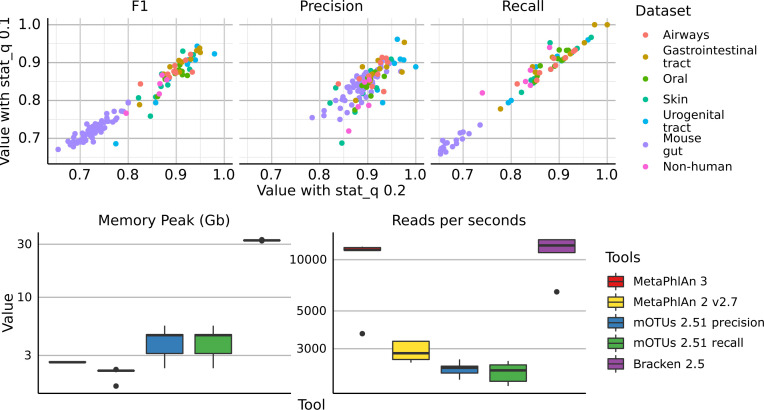
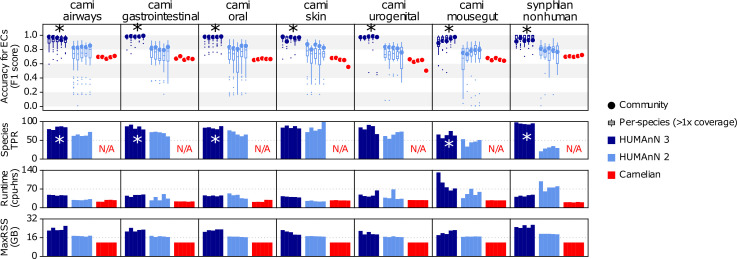
(A) The newly developed ChocoPhlAn 3 consolidates, quality controls, and annotates isolate-derived reference sequences to enable metagenomic profiling in subsequent bioBakery methods. (*The 1.1M MetaPhlAn 3 markers also encompass 61.8 k viral markers from MetaPhlAn 2 Truong et al., 2015) (B) MetaPhlAn 3 was applied to a set of 113 total evaluation datasets provided by CAMI (Fritz et al., 2019) representing diverse human-associated microbiomes and five datasets of non-human-associated microbiomes (Supplementary file 1). MetaPhlAn 3 showed increased performance compared with the previous version MetaPhlAn 2 (Truong et al., 2015), mOTUs2 (Milanese et al., 2019), and Bracken 2.5 (Lu et al., 2017). We report here the F1 scores (harmonic mean of the species-level precision and recall, see Figure 1—figure supplement 1 for other evaluation scores). (C) MetaPhlAn 3 better recapitulates relative abundance profiles both from human and murine gastrointestinal metagenomes as well from non-human-associated communities compared to the other currently available tools (full results in Figure 1—figure supplement 1). Bracken is reported both using its original estimates based on the fraction of reads assigned to each taxon and after re-normalizing them using the genome lengths of the taxa in the gold standard to match the taxa abundance estimate of the other tools. (D) Compared with HUMAnN 2 (Franzosa et al., 2018) and Carnelian (Nazeen et al., 2020), HUMAnN 3 produces more accurate estimates of EC abundances and displays a higher species true positive rate compared to HUMAnN 2. In panels B–D, an asterisk ('*') indicates that the bioBakery 3 method (MetaPhlAn 3 or HUMAnN 3) scored significantly better than all other methods (repeated paired t-tests over synthetic metagenomes, two-tailed p<0.05).