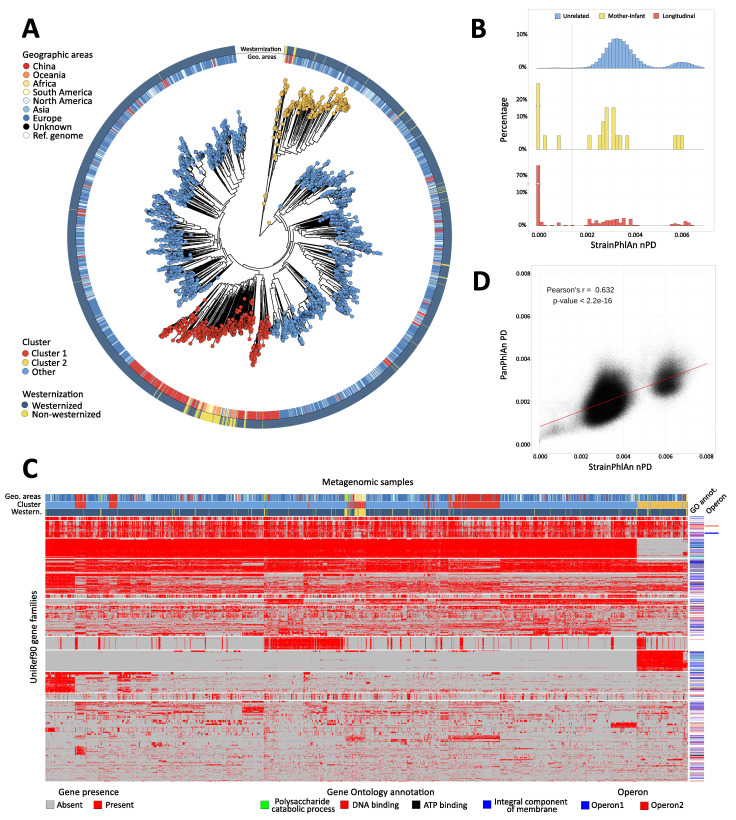
Figure 4. Population-scale strain-level phylogenetic and pangenomic analyses of Ruminococcus bromii from over 4000 human gut metagenomes.
(A) StrainPhlAn 3 profiling revealed stratification of Ruminococcus bromii clades with genetic content and variants frequently structured with respect to geographic origin and lifestyle. Genetically divergent subclades were identified, labeled as ‘Cluster 1’ (mainly composed of strains retrieved from Chinese subjects, Fisher’s exact test p<2.2e-16) and a subspecies-like Cluster 2. (B) Strain tracking of R. bromii. While unrelated individuals from diverse populations very rarely share highly genetically similar strains, pairs of related strains are readily detected by StrainPhlAn from longitudinal samples from the same individuals (quantifying short- and medium-term strain retention at about 75%) and in mother-infant pairs (confirming this species is at least partially vertically transmitted). Normalized phylogenetic distances (nPD) were computed on the StrainPhlAn tree. (p<0.003 two-sample Kolmogorov–Smirnov test on all the three distributions) (C) PanPhlAn 3 gene profiles of R. bromii strains from metagenomes highlights the variability and the structure of the accessory genes across datasets (core genes were removed for clarity). A total of 6151 UniRef90 gene families from the R. bromii pangenome were detected across the 2679 of the 4077 samples in which a strain of this species was present at a sufficient abundance to be profiled by PanPhlAn. The 13 highest-rooted gene clusters are shown, highlighting co-occurrence of blocks likely to be functionally related. The most common GO annotations are also reported together with two operons containing genes verified to be on the same locus by analysis of the reference genomes in the PanPhlAn 3 database. (D) Genetic (SNV on marker genes from StrainPhlAn 3) and genomic (gene presence/absence from PanPhlAn 3) distances between R. bromii strains are correlated (Pearson's r = 0.632, p-value<2.2e-16) pointing at generally consistent functional divergence in this species.