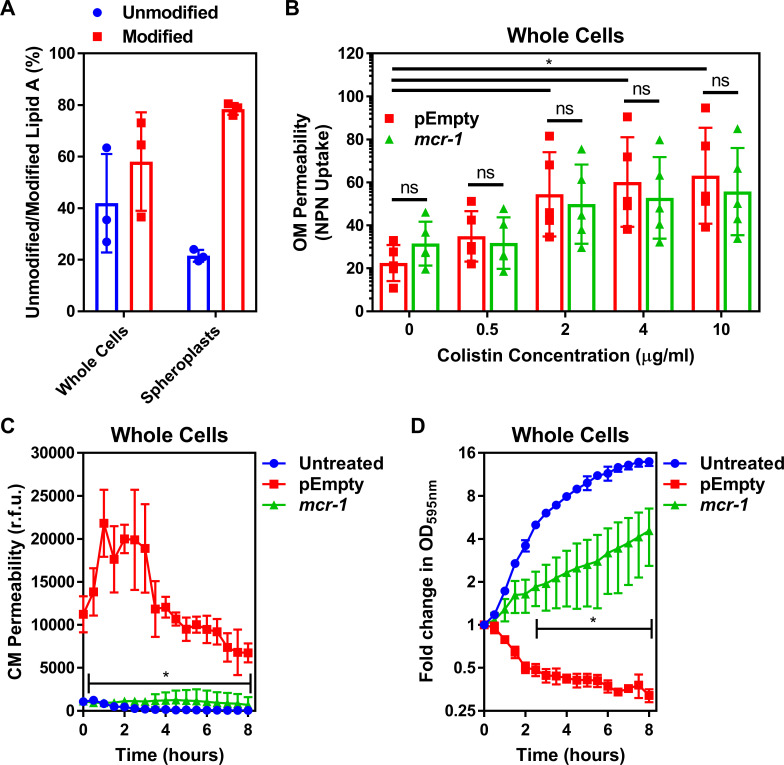
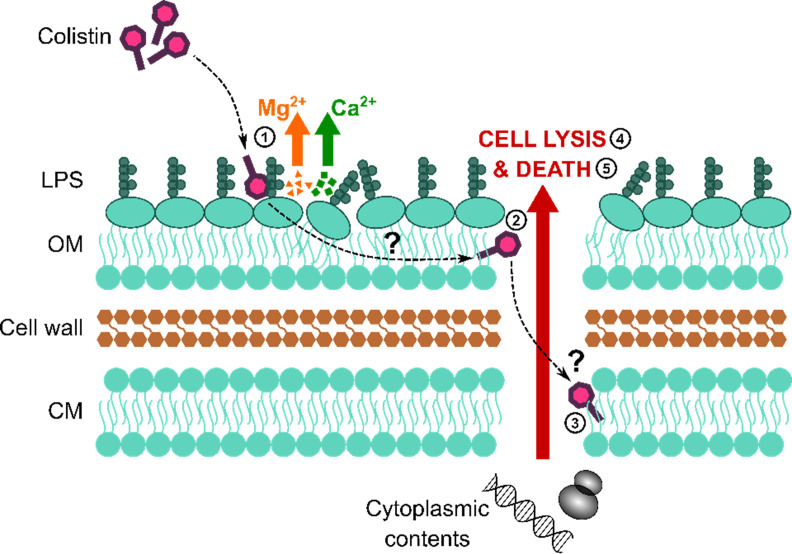
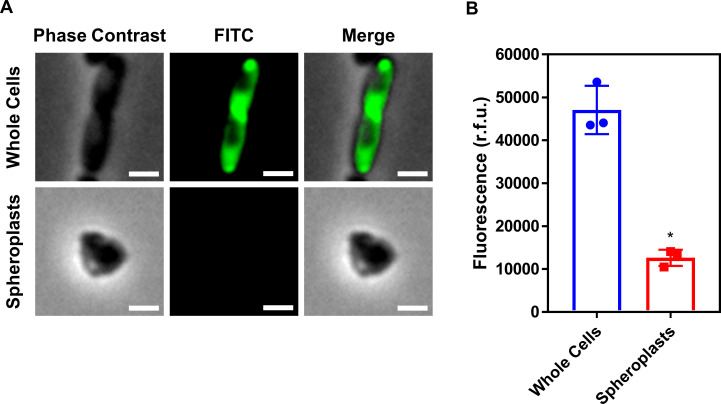
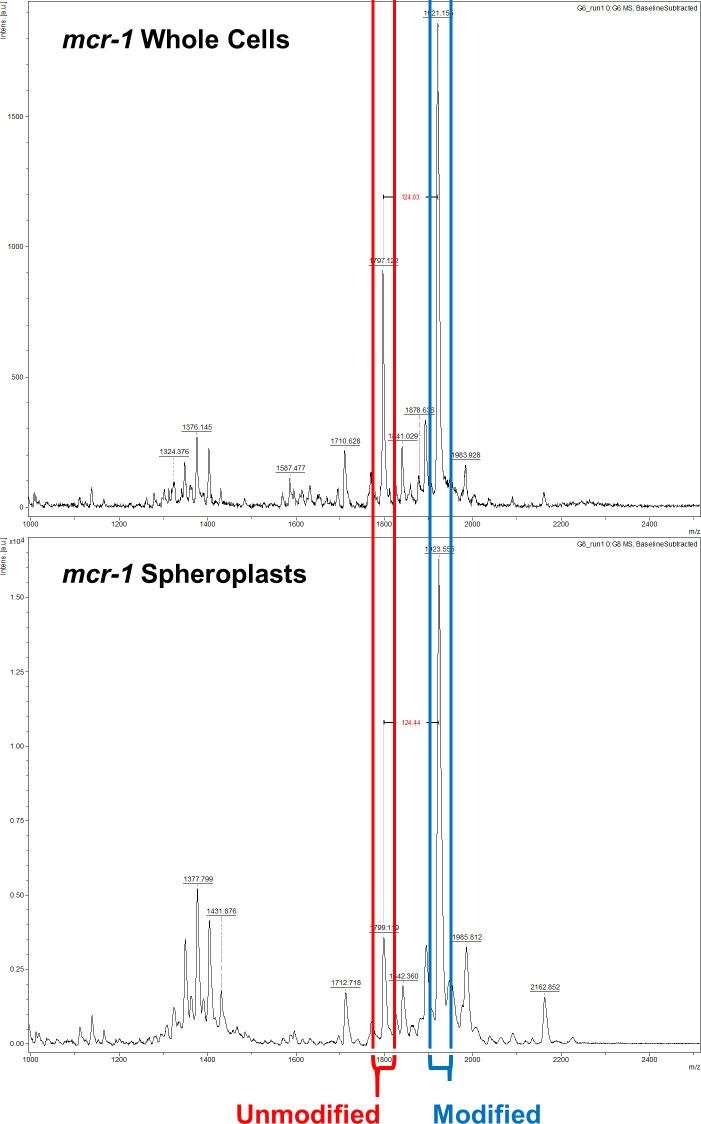
Figure 1. Colistin disrupts the outer membrane but not the cytoplasmic membrane of E. coli expressing mcr-1.
(A) Quantification of LPS modified with phosphoethanolamine, expressed as the percentage of unmodified lipid A and unmodified lipid A, in whole cells and spheroplasts of E. coli MC1000 expressing mcr-1, as determined by MALDI-TOF-based lipidomics (n = 3 in duplicate, *p<0.05 between Whole Cells and Spheroplasts). (B) OM disruption of E. coli MC1000 cells expressing mcr-1 or an empty plasmid control strain (pEmpty) during 10 min of exposure to colistin at the indicated antibiotic concentrations, as determined by uptake of the fluorescent dye NPN (10 µM) (n = 5, each data point represents the arithmetic mean of 20 replicate measurements; ns: p>0.05 between pEmpty and mcr-1 strains, *p<0.05 between the indicated concentrations of colistin). (C) Permeabilisation of the CM of E. coli MC1000 cells expressing mcr-1 or empty plasmid-containing cells during incubation with colistin (4 µg ml−1), as determined using 2.5 µM propidium iodide (PI) (n = 4; *p<0.0001 between pEmpty and mcr-1 strains). (D) Growth or lysis of E. coli MC1000 cells expressing mcr-1 or empty plasmid control cells during exposure to colistin (4 µg ml−1), as measured using OD595nm readings (n = 4; *p<0.05 between pEmpty and mcr-1 strains). Data in (A) were analysed by a two-tailed paired Student’s t-test. Data in (B–D) were analysed by a two-way ANOVA with Sidak’s (B) or Dunnett’s (C, D) post hoc tests. Data are presented as the arithmetic mean, and error bars represent the standard deviation of the mean. OM: outer membrane; NPN: N-phenyl-1-naphthylamine; CM: cytoplasmic membrane; r.f.u.: relative fluorescence units; OD: optical density.