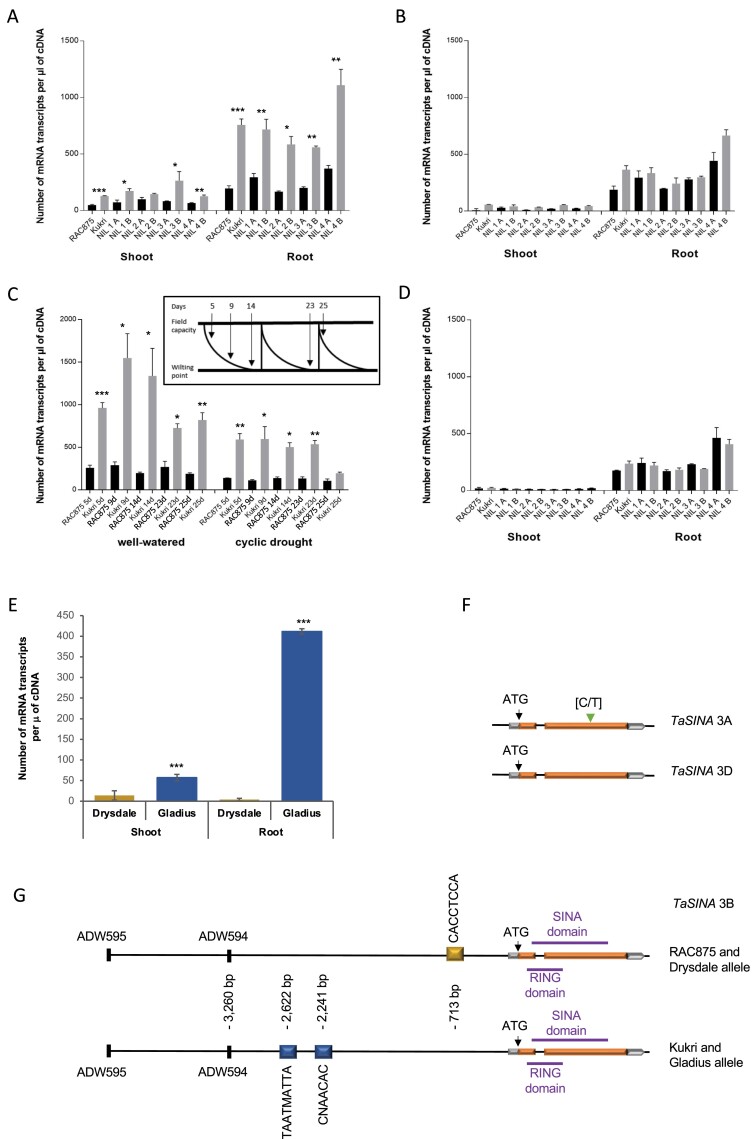
Fig. 5.
Expression analysis and gene structure of TaSINA and its homeologues. (A, E) Expression analysis of TaSINA in seedling shoot and root tissues of RAC875, Kukri, and NILs (A) and in Gladius and Drysdale (E). (C) Expression of TaSINA in RAC875 and Kukri under well-watered and cyclic drought conditions. (B, D) Expression analysis of TaSINA homeologues on chromosomes 3A (B) and 3D (D). ‘A’ corresponds to the RAC875 allele and ‘B’ corresponds to the Kukri allele. *P-value <0.05; **P-value <0.01; ***P-value <0.001. (F) Exon–intron structure of TaSINA homeologues on chromosomes 3A and 3D. (G) Exon–intron structure and promoter annotation of TaSINA on chromosome 3B. The CACCTCCA domain is a putative ABSCISIC ACID INSENSITIVE-4- (ABI4) binding site called S box (SBOXATRCS). Yellow boxes indicate the position of cis-acting elements present in the promoter region; green triangle, SNP position inducing the alanine to valine amino acid change; orange boxes represent the exons and the grey boxes the UTRs.