Fig. 3.
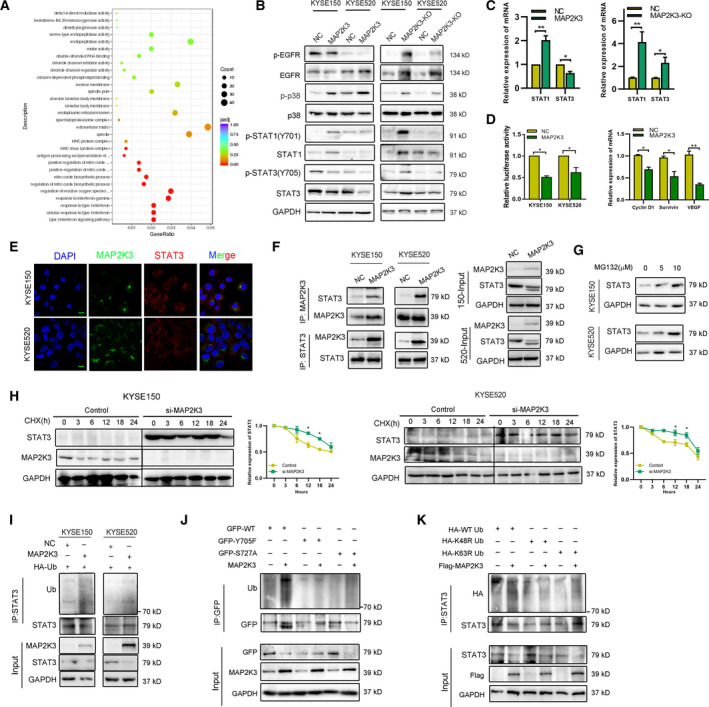
MAP2K3 modulates the EGFR/STAT3 signaling pathway in ESCC by promoting its proteasome degradation. (A) Distribution of the top 20 enriched GO terms in biology process, cellular component, and molecular function for the differentially expressed genes in MAP2K3‐overexpressing ESCC cells based on RNA‐seq analysis. (B) Immunoblot analysis to detect phosphorylation and total EGFR, p38, STAT1, and STAT3 protein expression in ESCC cells. (C) qPCR analysis to detect STAT1 and STAT3 RNA expression after MAP2K3 knockout and transfection in KYSE520 cells. (D) The transcription activity of STAT3 was detected after MAP2K3 transfection by luciferase reporter assay (left panel). The mRNA expression of STAT3 downstream genes was detected by qRT‐PCR (right panel). (E) Colocalization of STAT3 and MAP2K3 was detected by immunofluorescence in KYSE150 and KYSE520 (scale bar: 20 µm). (F) Binding of endogenous MAP2K3 with STAT3 was detected by co‐immunoprecipitation in KYSE150 and KYSE520 cells. (G) Expression of STAT3 was detected by western blot after different doses of MG132 treatment for 24 h. (H) ESCC cells were treated by cycloheximide (CHX, 200 µg·mL−1) in a time‐dependent manner after transfecting si‐MAP2K3 and control. I. STAT3 ubiquitination was detected after MAP2K3 transfection by immunoprecipitation with anti‐STAT3 antibody and immunoblotting with anti‐Ub. (J) GFP‐STAT3 (WT), GFP‐STAT3 (Y705F), or GFP‐STAT3 (S727A) were transfected into KYSE150 cells together with the MAP2K3 plasmid or control, then STAT3 ubiquitination was detected. (K) HA‐tagged wild‐type, K48R, and K63R Ub were transfected into KYSE150 cells together with the MAP2K3 plasmid. STAT3 ubiquitination was detected. Error bars represent the SD from at least three independent biological replicates. (*P < 0.05; **P < 0.01; ***P < 0.001 by Student’s t‐test).
