Fig. 5.
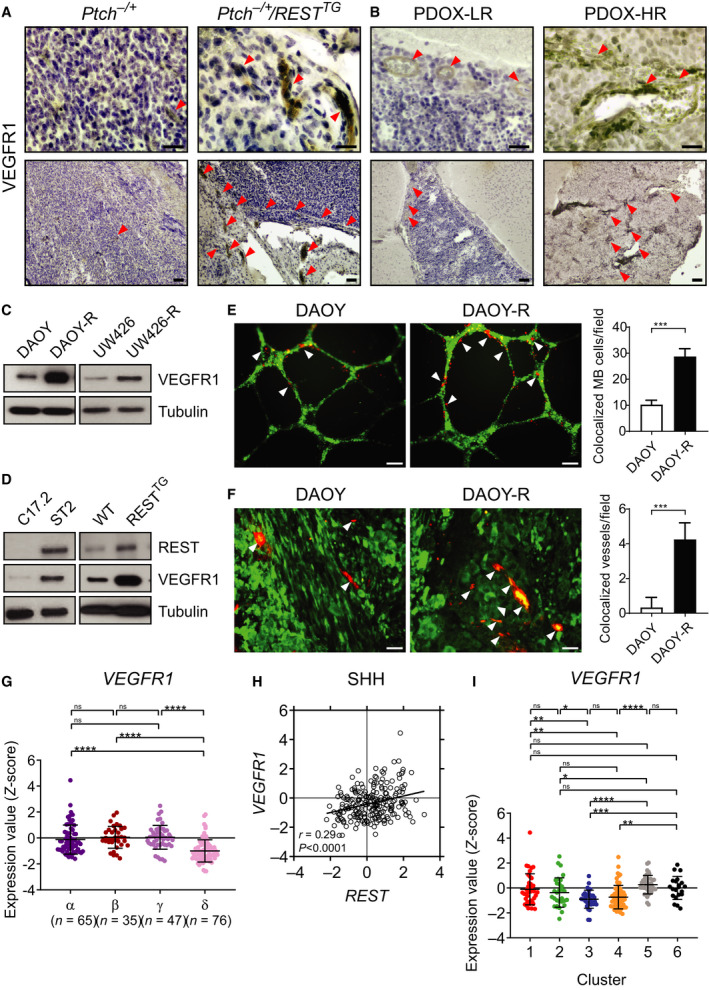
MBs with elevated REST expression upregulate VEGFR1 and colocalize with endothelial cells. (A‐B) IHC was performed on cerebellar sections (n = 3) from Ptch+/− and Ptch+/−/RESTTG mice and PDOX animals (n = 3) to demonstrate VEGFR1 expression. Arrowheads show blood vessels; Scale bars: top (40×) = 10 μm, bottom (10×) = 20 μm. (C‐D) Western blot analysis to measure VEGFR1 levels in human DAOY/DAOY‐R and UW426/UW426‐R cells, and VEGFR1 and REST protein levels in mouse C17.2/ST2 cells, and in CGNPs from WT/RESTTG mice. Tubulin served as a loading control. (E) HUVECs were cocultured with DAOY or DAOY‐R cells on matrigel, and tube formation was assessed after 16 h. DAOY/DAOY‐R are in red, while HUVECs are shown in green color. Quantitation of the relative numbers of DAOY or DAOY‐R cells colocalized with HUVECs is shown on the right. (F) Immunofluorescence assay to show colocalization in yellow of CD31‐positive endothelial cells (red) and luciferase‐positive tumor cells (green) in tumor sections from DAOY or DAOY‐R xenografts. Quantitative data (n = 3, three fields/section) is shown on the right. P‐values were obtained using Student's t‐test. ***P < 0.001. (G) Profile of VEGFR1 mRNA expression in microarray data of four subtypes of human SHH‐MB samples from GSE85217 data set [4]. Each dot corresponds to one individual patient. Data show individual variability and means ± SD. P‐values were obtained using the unpaired t‐test with Welch's correction. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. (H) Scatter plot of correlation of REST mRNA expression and VEGFR1 mRNA expression. Figure shows the plot across all 223 SHH‐MB patients (r = 0.29, P < 0.0001). (I) VEGFR1 mRNA expression profile in SHH‐MB patient samples. Hierarchical clustering based on expression of neuronal differentiation markers divided the SHH‐MB patient samples into six distinct clusters [10]. Each dot represents an individual patient. Data show individual variability and means ± SD. P‐values were obtained using the unpaired t‐test with Welch's correction. ns, not significant. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
