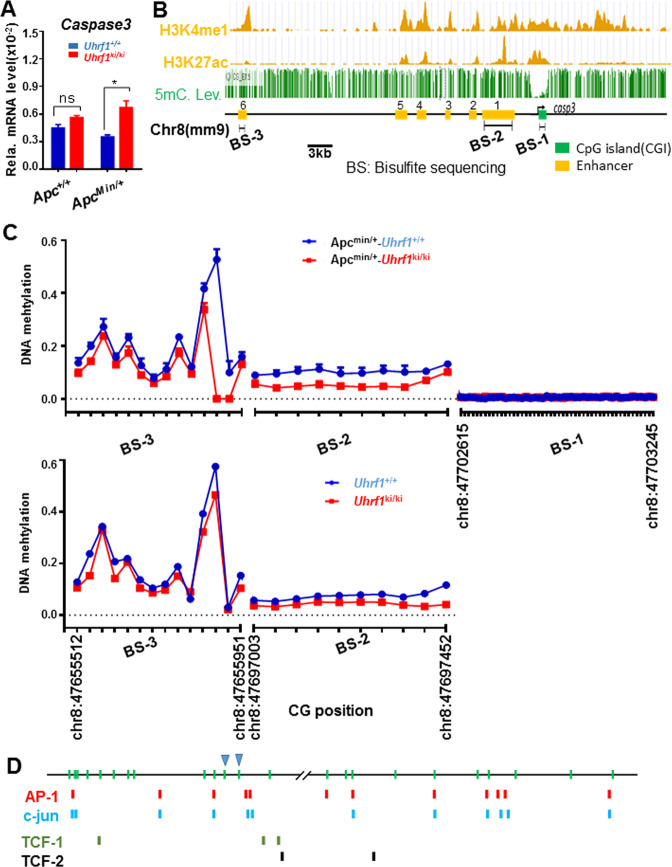
Fig. 5. Activation of caspase-3 gene expression in Uhrf1ki/ki/Apcmin/+ mice correlates with reduced DNA methylation in the caspase-3 enhancer regions.
A qRT-PCR analysis showing elevated levels of caspase-3 mRNA in small intestinal epithelium cells from Uhrf1ki/ki/Apcmin/+ mice. The levels of caspase-3 mRNA were determined from three pairs of littermate mice and were normalized to Actin. *p < 0.05; ns, p > 0.05. Three mice were analyzed for each genotypes. Error bars, S.E. B Diagrams illustrating the epigenetic marks H3K4me1 and H3K27ac landscapes at the mouse caspase-3 gene. The profiles were derived from small intestinal tissue ChIP-seq data in UCSC genome browser. The putative enhancers, from enhancer 1 to 6, were assigned according to overlapping H3K27ac and H3K4me1 peaks. As a reference, also shown are the DNA-methylation status within or upstream of casepase-3 gene derived from E7.5 mouse embryos (downloaded from Methbank database). C Summary of bisulfite sequencing results of DNA-methylation status within promoter, enhancer 1 and enhancer 6 regions. Each CpG sites were sequenced over hundred or thousand times by high-throughput Illumina® (ND102-0102) sequencing and thus the data were highly accurate. Note that the caspase-3 promoter region in small intestinal epithelium cells is essentially unmethylated in both Uhrf1+/+/Apcmin/+ and Uhrf1ki/ki/Apcmin/+ mice. D Caspase-3 enhancer 1 and enhancer 6 contain multiple binding sites for Wnt-regulated transcription factors TCF1/2, AP-1, and c-jun. The two highly demethylated CpG sites are marked with triangle.