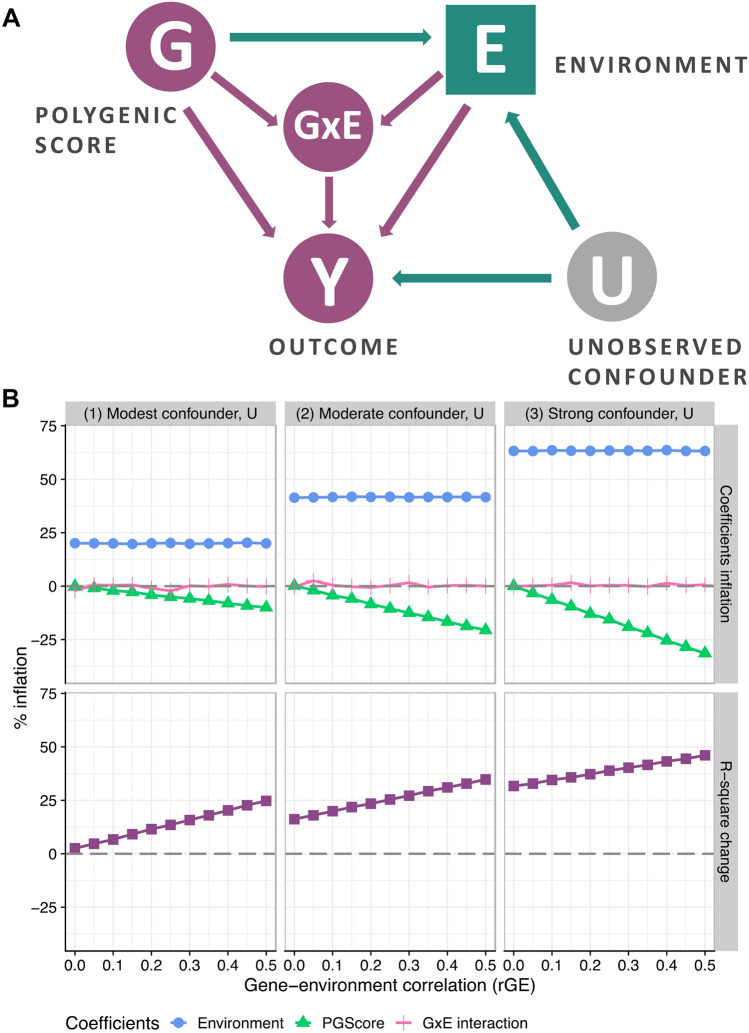
Figure 2.
Collider bias in polygenic gene-environment interaction models. Panel A. Schematic diagram of the collider bias which occurs between polygenic score, environment, and outcome in cases of gene-environment interdependence. Dark purple circles represent variables, unobserved confounders are shown in grey circles, collider variables are indicated in squares. By adding E into the model with the polygenic score G, we make E a collider. A collider that is not conditioned on, blocks the path between its sources (G and U); once a collider is controlled for, the path is opened as indicated by green nodes. Panel B (top). Spurious regression estimates for the polygenic score and environment along with non-inflated interaction terms from the series of OLS simulations reflecting a range of gene-environment interdependence and the presence of modest, moderate, or strong confounder, U. Collider bias due to positive values of gene-environment correlation and the presence of an uncontrolled confounder, which is positively correlated with covariate and outcome, results in deflation of polygenic score estimates. Deflation is greater the higher the gene-environment correlation; greater confounding also results in greater bias. The interaction term is not affected but results for moderation analysis are biased as long as direct effects are spurious. Panel B (bottom). R-squared inflation plot from the series of OLS simulations; collider bias results in inflated values of explained variance statistics. R-squared statistics for the model with endogenous covariate and polygenic score includes not only the true share of the variance in Y explained by G and E (baseline estimate indicated by 0), but also the elements of variance that are due to gene-environment correlation and confounder(s), U.