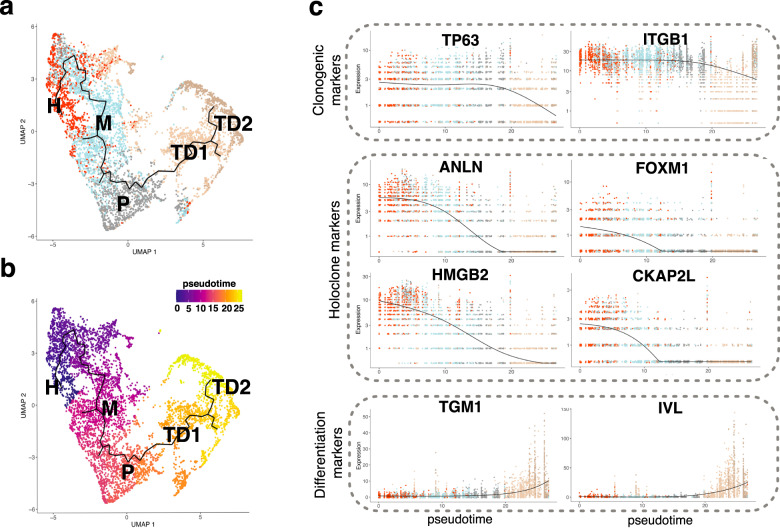
Fig. 3. Single-cell RNA-seq analysis identifies a unique trajectory among the clusters.
a UMAP plot of cells annotated as keratinocytes identified at t1 displaying the result of Monocle3 analysis. The identified trajectory is shown as a continuous black line. Cells are colored according to cluster identity determined with Seurat analysis. b Same as a, cells are colored according to pseudotime. c Kinetics plot showing expression values normalized and rounded by Monocle3 of two representative clonogenic markers (TP63, ITGB1), four representative markers present in the Holoclone signature (ANLN, FOXM1, HMGB2, CKAP2L) and two representative differentiation marker (TGM1 and IVL) genes across pseudotime.