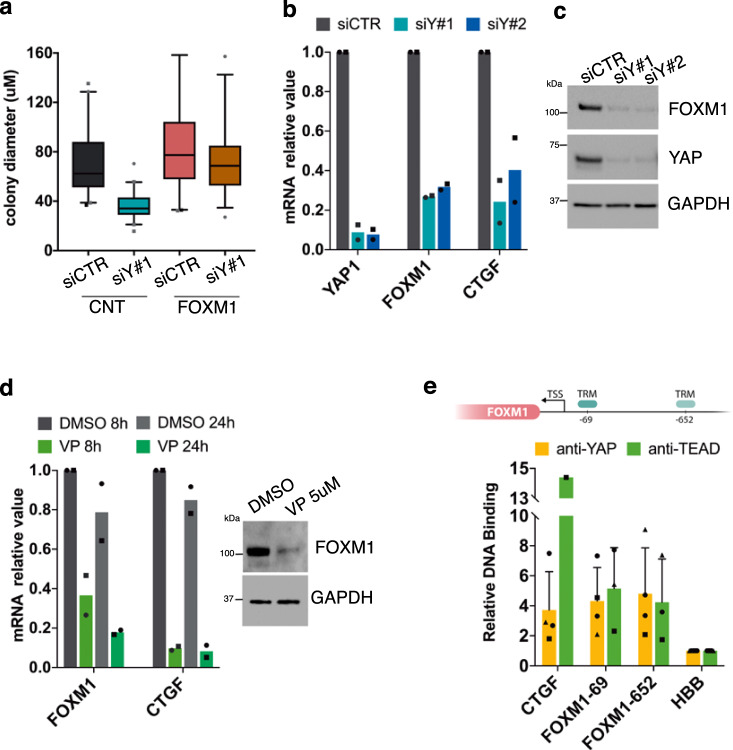
Fig. 5. FOXM1 acts downstream of YAP.
a Box plot showing the diameter of colonies stained after 5 days of cultivation of CNT or FOXM1-transduced keratinocytes both transfected with control (siCTR) and YAP-specific (siY#1) siRNA. Data from one representative experiment. Enforced FOXM1 rescued the colony growth potential impaired by downregulation of YAP. Median and 5–95 percentile displayed. b qRT-PCR on mRNAs obtained from keratinocytes transfected with control (siCTR) and YAP-specific (siY#1, siY#2) siRNA. Expression levels of YAP1, FOXM1, and CTGF were normalized per GAPDH and given relative to siCTR (arbitrarily set to 1) (n = 2 biological replicates derived from independent human primary keratinocyte cultures, indicated with dots and squares, respectively). c Western analysis of total cell extracts from cultures treated with the indicated siRNA shows that FOXM1 expression is almost abolished after YAP depletion (representative images of n = 3). d left: qRT-PCR on mRNAs obtained from keratinocytes treated with Verteporfin (VP 5 μM) or vehicle (DMSO) for the indicated time. Expression levels of FOXM1 and CTGF were normalized per GAPDH and given relative to DMSO, 8 h (arbitrarily set to 1) (n = 2 biological replicates derived from independent human primary keratinocyte cultures, indicated with dots and squares, respectively). Right: Western analysis of total cell extracts from cultures treated with DMSO or VP 5 μM show that FOXM1 protein decreases after 24 h of treatment. e Up: Scheme showing two TEAD-recognition motifs (TRM), respectively, at −69 (ref. 35) and −652 nucleotide36 from transcription start site (TSS) into the FOXM1 promotorial region. Down: ChIP-qPCR showing YAP binding to the indicated sites in human primary keratinocytes. Relative DNA binding was calculated as a fraction of input and given relative to the negative region (HBB) arbitrarily set to 1; average and standard deviation displayed from four independent biological replicates, indicated with different shapes.