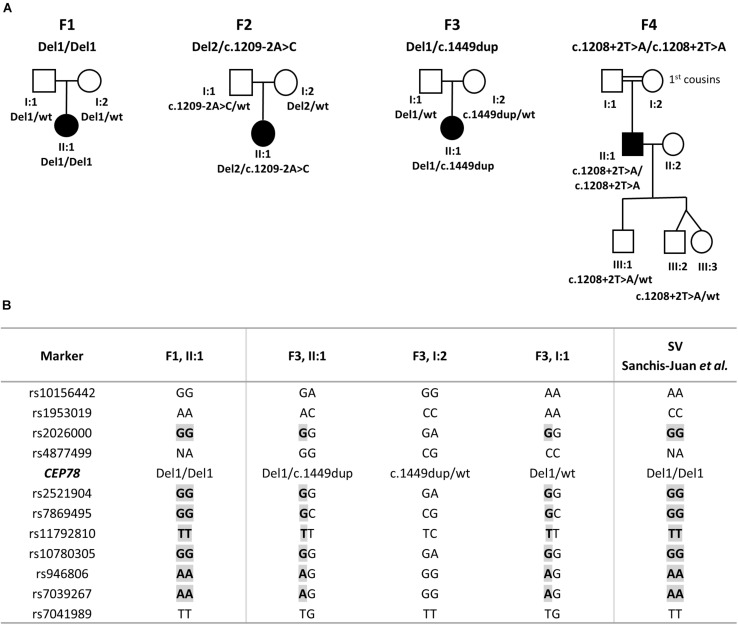
FIGURE 1.
Pedigrees segregating the CEP78 structural and sequence variants and haplotype reconstruction for the shared SV. (A) F1, II:1 is homozygous for a complex deletion–inversion–deletion involving exons 1–5 of CEP78. F2, II:1 is compound heterozygous for a deletion of 235 kb overlapping CEP78 and PSAT1 and for splice site variant c.1209-2A>C. F3, II:1 is compound heterozygous for the same deletion–inversion–deletion of F1, II:1 and the c.1449dup variant in CEP78. F4, II:1 is homozygous for splice site variant c.1208+2T>A. Extended F4 pedigree available in Supplementary Clinical Data 2. (B) Genotyping of 18 flanking single-nucleotide polymorphisms (SNPs) revealed a common haplotype of 1.9 Mb, between the individuals carrying the deletion–inversion–deletion [F1, F3 and the case originally described by Sanchis-Juan et al. (2018)]. Extended version available in Supplementary Table 5. Abbreviations: Del, deletion (or deletion–inversion–deletion); wt, wild type.