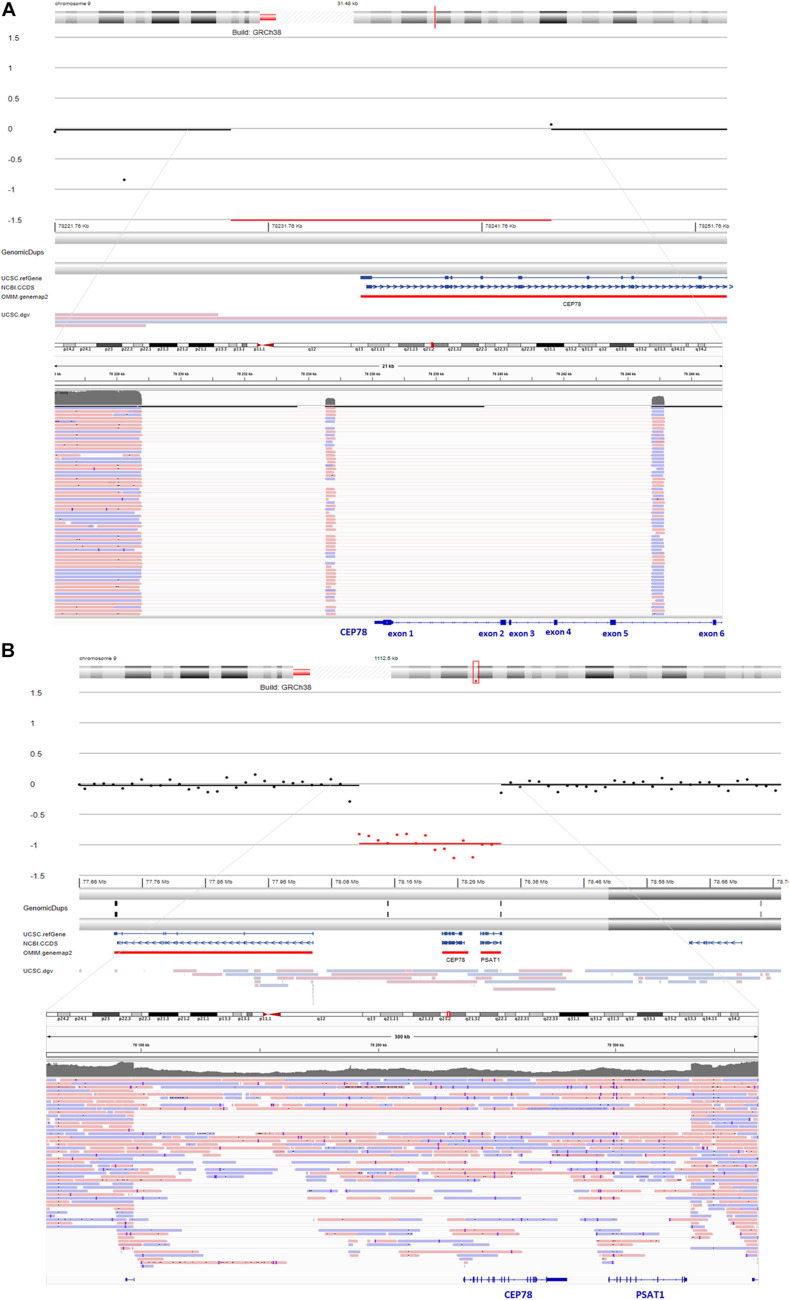
FIGURE 2.
sWGS output and integrative genomics viewer (IGV) view of the structural variants (SVs) detected via long-read sequencing (LRS) in F1 and F2. (A) sWGS in F1, II:1 revealed a homozygous deletion of the region spanning exons 1–5 of CEP78 (top). Subsequent delineation of the SV via targeted LRS of the junction product identified a complex deletion–inversion–deletion (bottom). The left breakpoint is located at (hg38) chr9:78228782, whereas the right breakpoint is located at chr9:78244762. The inverted segment spans chr9:78234546–78234844 [nearby an L1ME3Cz repetitive element (chr9:78234521–78234902)]. The SV overlaps with a previously described SV affecting CEP78 (Sanchis-Juan et al., 2018). (B) sWGS in F2, II:1 identified a heterozygous deletion spanning the entire CEP78 and PSAT1 genes (top). Final delineation was obtained via whole-genome LRS, coordinates are crossing chr9: 78096930–78331887 and cover 235 kb (bottom).