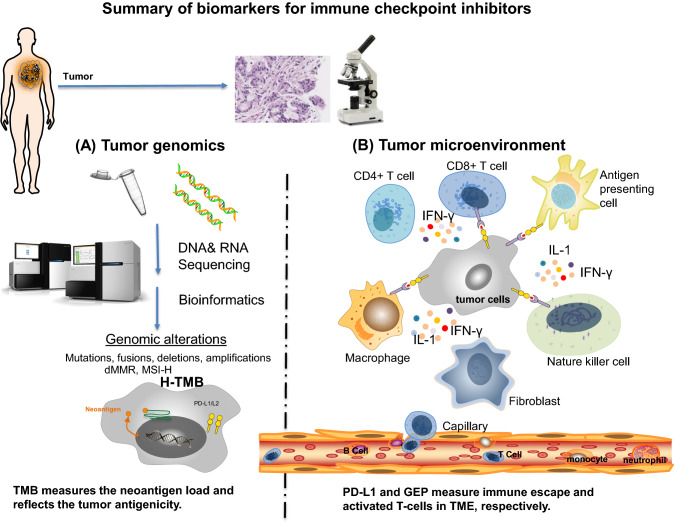
Fig. 1.
Summary of biomarkers for immune checkpoint inhibitors. The tumor microenvironment can be examined via histopathology and molecular studies. The relationship between tumor and immune cells can be evaluated by immunohistochemistry and molecular or genetic profiling analysis. Known biomarkers for immune checkpoint inhibitors characterize the properties of either tumor genomics (a) or tumor microenvironment (b). a Next generation sequencing (NGS) and bioinformatics identify genomic alterations in tumor cells, which include somatic mutations, fusions, deletions, amplifications, dMMR, MSI-H and H-TMB. Among these genomic biomarkers, TMB best measures the neoantigen load and reflects the tumor antigenicity. b TME includes tumor cells and its surrounding blood vessels, fibroblasts, immune cells (e.g., lymphocytes), bone marrow-derived suppressed cells, extracellular matrix (ECM), and signaling molecules (e.g., interleukin (IL)-1, interferon-gamma (IFN-γ). Tumor cells escape immune surveillance via PD-L1 expression. The combination of PD-L1 IHC and GEP better characterizes the immune escape and immune cell activity in TME. dMMR deficient mismatch repair; H-TMB high tumor mutation burden; MSI-H microsatellite instability high; GEP gene expression profiling; PD-L1 programmed death-ligand 1; IHC immunohistochemistry