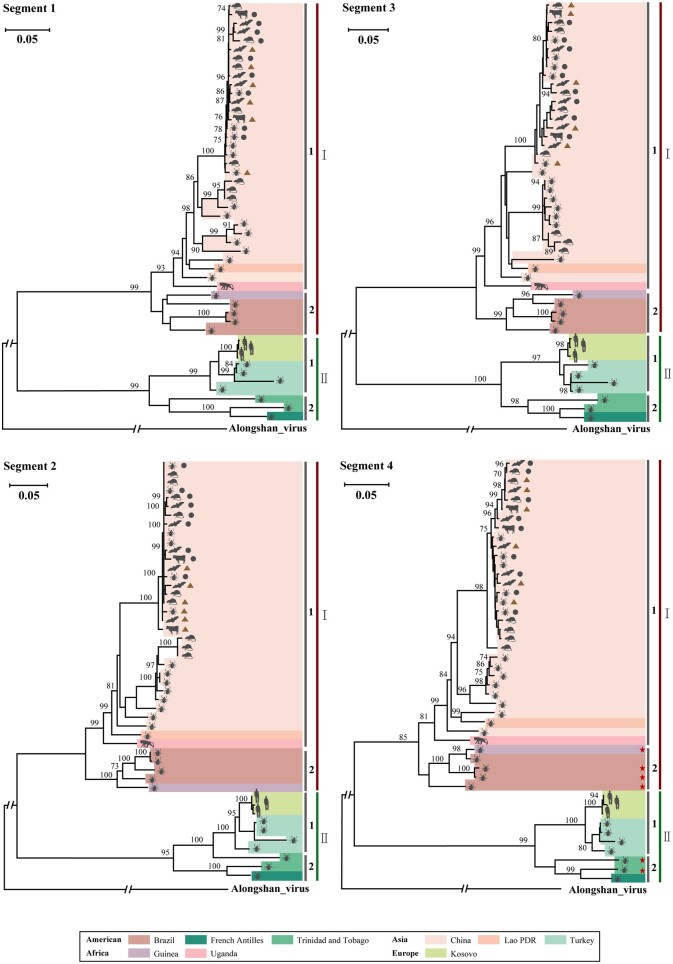
Figure 3.
Phylogenetic analysis of the nt sequences of four segments of JMTV. Because of the high sequence similarity of the JMTVs discovered in this study, representative sequences including six complete genomes (brown triangles) and nine nearly complete genomes (gray circles) were used to infer the evolutionary trees. The JMTVs containing three or four segments and available on GenBank were used as reference sequences. Phylogenetic groups, as well as host and geographical origin are indicated. The JMTV whose segment 4 encoded non–overlapping ORFs is marked with red stars in the segment 4 phylogenetic. All trees were rooted using Alongshan virus. Bootstrap values (>70%) are shown at relevant nodes. The scale bar represents the number of nt substitutions per site. The GenBank accession numbers of the viruses used in the trees are found in Supplementary Fig. 2.