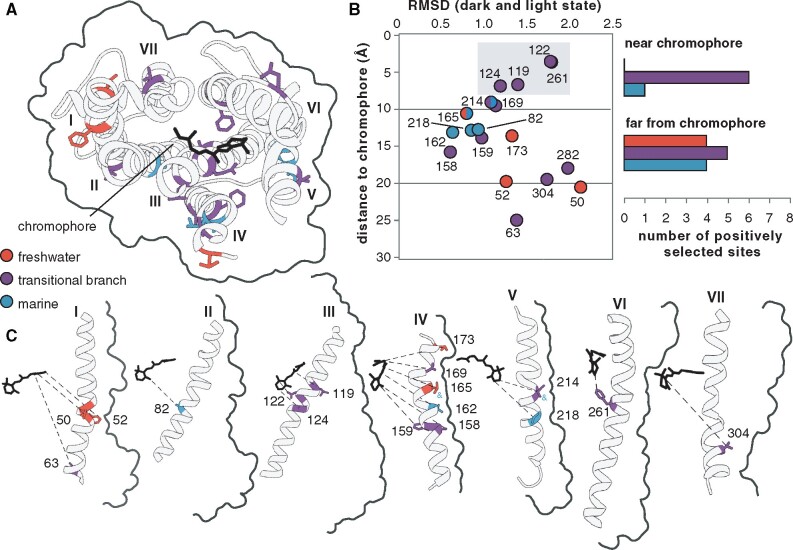
Fig. 3.
Habitat specificity of positively selected sites in croaker rhodopsin. (A) Positively selected sites on transmembrane helices shown on the rhodopsin dark-state crystal structure (1U19) looking down from the intradiscal face. (B) Distance from the chromophore and root mean square deviation between the dark (1U19) and light (3PQR) state rhodopsin crystal structures for each positively selected site. Histograms show the number of positively selected sites in each partition within ten angstroms of the chromophore where spectral tuning sites have been observed (Bowmaker and Hunt 2006). (C) Positively selected sites on each transmembrane helix shown arranged in order with the chromophore to the left. Sites under positive selection in more than one partition are indicated by an ampersand of the same color as the additional partition.