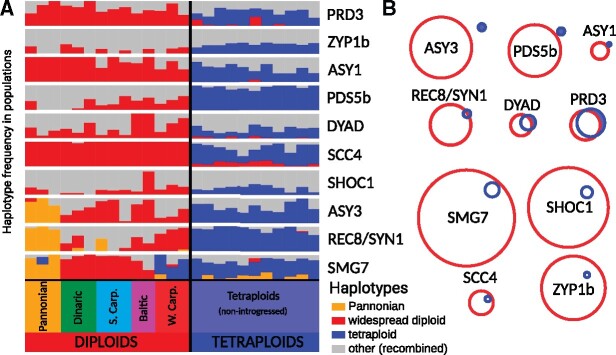
Fig. 2.
Limited standing variation across Arabidopsis arenosa diploids in protein candidates for tetraploid meiotic adaptation. (A) Lack of tetraploid-specific haplotypes in diploid populations sampled across the total range of A. arenosa. Haplotypes were combined across linked candidate AASs within each protein. A set of bar plots for each of ten candidate proteins (horizontal lines) shows frequencies of diploid, Pannonian (if different from widespread diploid) and tetraploid-specific haplotypes (y axis) in each of 14 diploid and 11 tetraploid populations (x axis, grouped to lineages and ploidies). Frequencies of minor frequency haplotypes found in either or both ploidies are summed in a gray column. (B) A hypothetical maximal variation among haplotypes of meiosis proteins in diploids and tetraploids, quantified by Hamming distances. The diameter of the red and blue circles denotes the full range of potential variability of haplotypes reconstructed by all combinations of AASs among all diploid and tetraploid individuals, respectively. The relative distance of the red and blue circles denotes the genetic distance between the diploid and tetraploid haplotypes. Overlap of both circles suggests that it is plausible that the tetraploid haplotype could have existed within the observed variation in diploids, even if the exact tetraploid haplotype was not found in our diploid sampling. Filled area of the tetraploid circle, nonoverlapping with diploid, represents the tetraploid haplotype space that cannot be explained by, and would not be expected to exist, within extant diploid AAS variation. The upper six proteins show evidence that their tetraploid haplotypes most likely accumulated additional mutations after diploid/tetraploid divergence.