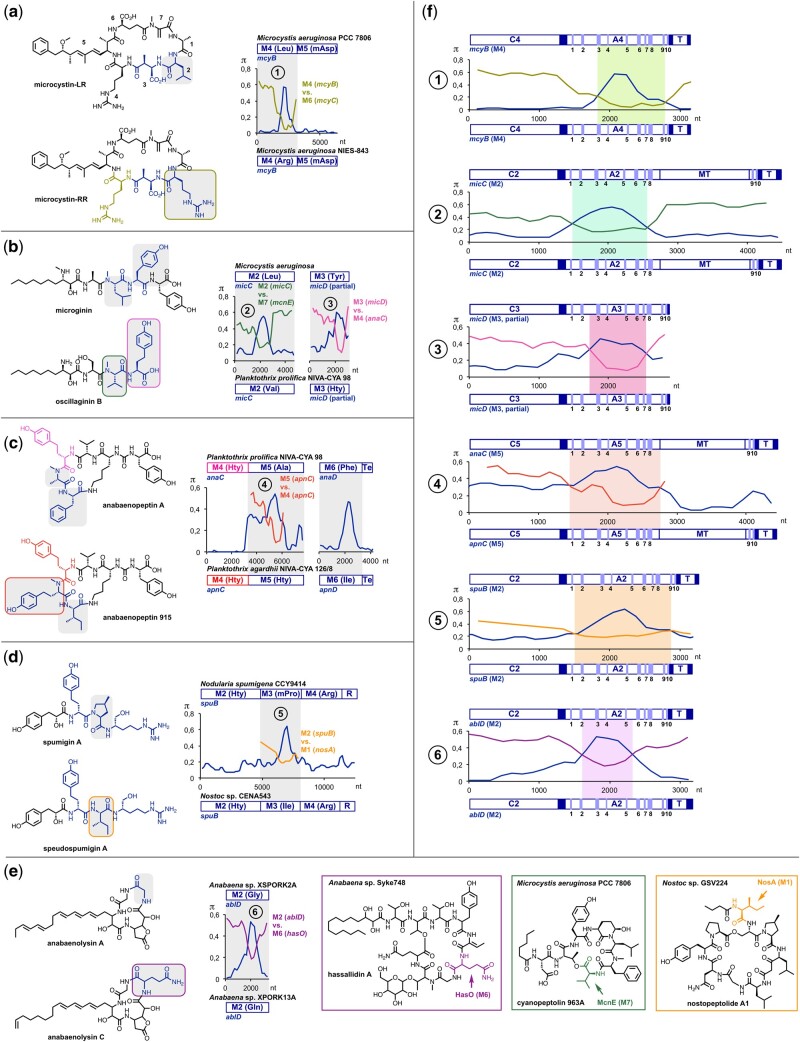
Fig. 2.
Diversification of cyanobacterial NRPs via recombination in the biosynthesis of (a) microcystins, (b) microginins, (c) anabaenopeptins, (d) spumigins, and (e) anabaenolysins. Structural differences between pairs from compound families (gray squares) correlate with nucleotide sequence divergence of the genes encoding NRPS modules (M). Related sequences have been aligned for pairwise comparison. π values (average number of nucleotide differences per site between two sequences) were computed using the sliding window mode in DnaSP (width, 300 nt; step, 150 nt). The mosaic structure of the genes (Smith 1992) clearly indicates recombination. This notion is also strongly supported by the detection of gene segments that complement divergent sites in a reciprocal fashion (numbered bullet points [BP] 1–6). Notably, the complement sequences stem from modules of the same cluster (BP 1, 4), from different clusters of the same species (BP 3), or from different clusters of different species (BP 2, 5, 6). Amino acid residues in the structures are color-coded to trace back their biosynthetic origin to individual modules. Hty, homotyrosine; Hph, homophenylalanine; mPro, 4-methylproline; mAsp, 3-methylaspartic acid; Te, thioesterase, R, reductive domain. (f) Close-up representation of putative recombination events to evaluate exchange unit boundaries. Gene segments encoding modules are divided into adenylation (A), condensation (C), thiolation (T), and, if present, methylation (MT) domains. Adenylation domain-specific core motifs are indicated by bands and numbers (1–10) (Marahiel et al. 1997). Linkers are indicated as filled squares. Highlighted parts of the graphs represent regions that are more closely related to sequences encoding other modules than to sequence of the respective ortholog.