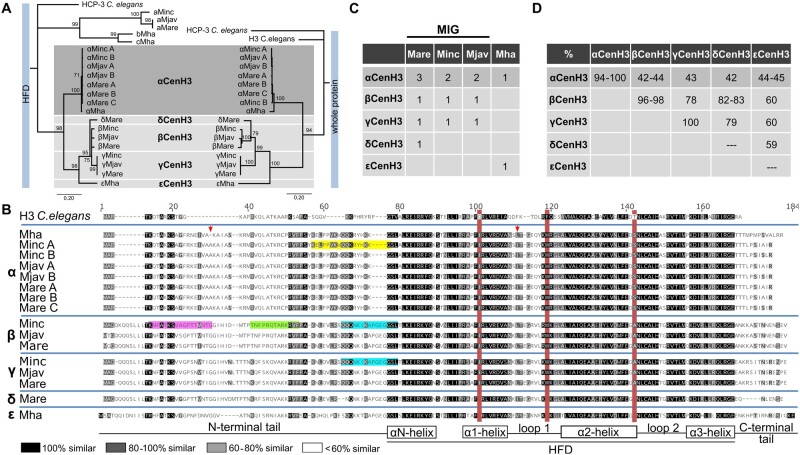
Fig. 1.
Identification of CenH3 proteins in Meloidogyne species (Minc, M. incognita; Mare, M. arenaria; Mjav, M. javanica; and Mha, M. hapla). (A) Phylogenetic analyses using NJ with a protein alignment of the (HFD) (left) and full length of CenH3 sequences (right) of all detected Meloidogyne CenH3s. Bootstrap values above 50 are displayed. CenH3 sequences are separated in the subgroups (a, b, c, α, β, γ, δ, and ε). (B) Amino acid alignment of CenH3 proteins separated into α, β, γ, δ, and ε subgroups. The red boxes indicate diagnostic amino acid changes in comparison to H3 from Caenorhabditis elegans. Secondary structure of histone-fold domain (HFD) is depicted below the alignment. The arrows indicate amino acids changes in αCenH3 from M. hapla in comparison to other αCenH3s. Highlighted are peptide sequence regions which were used as antigens to produce antibodies to αCenH3 (yellow), βCenH3-1 (green), βCenH3-2 (magenta), and βγCenH3 (cyan). (C) Copy number of CenH3 genes in Meloidogyne genomes. (D) Sequence identity matrix of CenH3 protein sequences.