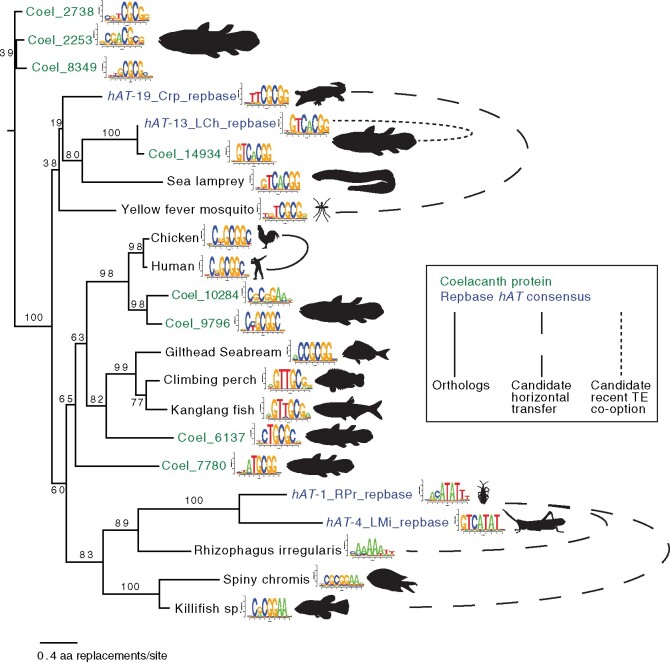
Fig. 2.
Phylogram of CGGBPs with DNA-binding motif displayed. Proteins with DNA-binding preferences on PBMs were aligned with Mafft E-INSi v7.310 (Katoh and Standley 2013), and any positions with gaps in greater than 20% of the sequences were trimmed using TrimAI. The unrooted maximum likelihood (ML) tree was determined using RAxML-NG (Kozlov et al. 2019), using the LG substitution matrix, ML estimated invariant site proportions, and a four-category gamma model of rate heterogeneity (LG+I+G4), and 300 bootstrap trees. The substitution model was selected according to maximum AIC as determined by ModelTest-NG. Motifs were produced using Top10AlignZ (Weirauch et al. 2014).