Figure EV5. Endothelial cells from human IGF‐1 receptor endothelial overexpressing mice (hIGFREO) can communicate with the gut wall.
-
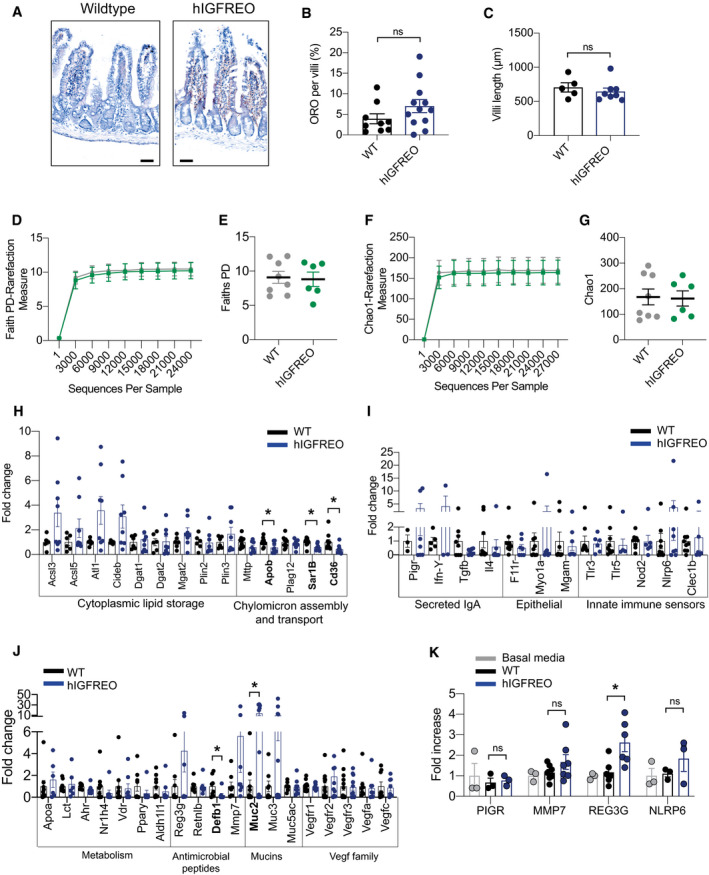
A–C(A), Representative images of haematoxylin‐ and eosin‐stained villi from hIGFREO compared with WT after 8 weeks of HFD. No difference in villi lipid content (B) or villi length (C) in hIGFREO compared with WT (n = 5–12 mice per group).
-
D, EFaith’s phylogenetic diversity (PD) was used to measure the faecal microbial diversity and demonstrates no difference between hIGFREO mice and WT on chow diet (n = 5‐8 mice per group).
-
F, GChao‐1 analysis was used to measure the faecal microbial diversity and abundance and again demonstrates no difference between hIGFREO and WT mice on chow diet (n = 5–8 mice per group).
-
H–JTargeted gene expression of the small intestine from hIGFREO normalised to WT after 8 weeks of high‐fat diet demonstrating a significant increment in Muc2 and decrement in Cd36, Sar1b, Apob and Defb1 (n = 6–8 mice per group).
-
KGene expression of Caco‐2 cells treated with conditioned media from hIGFREO endothelial cells showed a significant increase in Reg3g expression (n = 3–6 mice per group).
Data information: Data shown as mean ± SEM and individual mice are shown as data points, P < 0.05 taken as being statistically significant using Student’s t‐test and denoted as *. Ns denotes not significant. Diversity analyses were run on the resulting OTU/feature.biom tables to provide both phylogenetic and non‐phylogenetic metrics of alpha and beta diversity. Additional data analysis (PLS‐DA) and statistics were performed with R.
