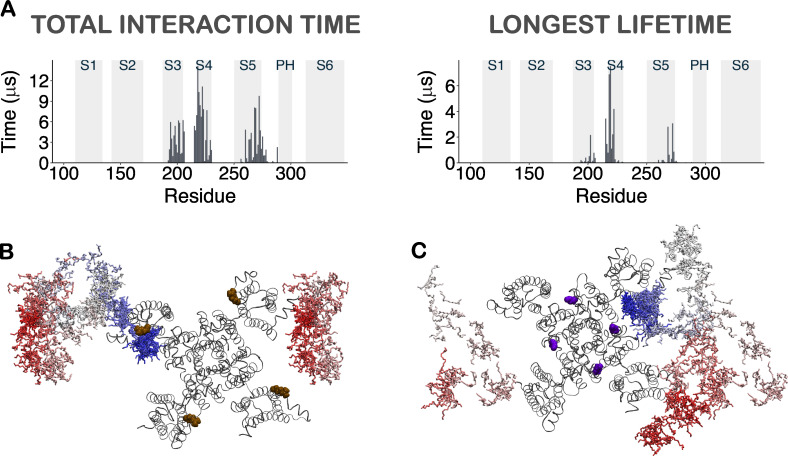
Figure S5.
Pattern of LIN interaction with KCNQ1 in AA simulations is preserved in longer and initial seed unbiased simulations. (A) To assess whether the AA simulations we performed have allowed sufficient sampling of binding events, we continued the LIN site I system for an additional 10 µs for better sampling of the interaction sites. Contact analysis for the 15-µs AA LIN site I simulation. Histogram of total interaction time (left) and longest lifetime (right) of each residue in the transmembrane segment of KCNQ1 for LIN. The gray highlighted boxes in the background indicate the position of the transmembrane helices. The contact analysis performed for the entire 15-µs AA simulation follows the exact same trend as the initial 5 µs with the same identified constant binders keeping their close interaction with the channel. (B and C) To evaluate and rule out any potential convergence/initial seed system bias in AA MD simulations, a system in which no LIN molecules were in close proximity to sites I and II in the CG simulations, called LIN-dissociated, was selected as a seed system for one production run. In this control AA MD simulation, LIN molecules were allowed to diffuse in the multicomponent membrane for their preferred interaction sites. In 5-µs AA MD simulation started from LIN-dissociated state, LIN stably binds to each of the identified sites I and II. The path of a LIN molecule finding its way to site I (B) and site II (C) in the dissociated system, with red designating the starting point and blue marking the end point during a 5-µs simulation. Residues R218 and K316 are highlighted in brown and purple, respectively.