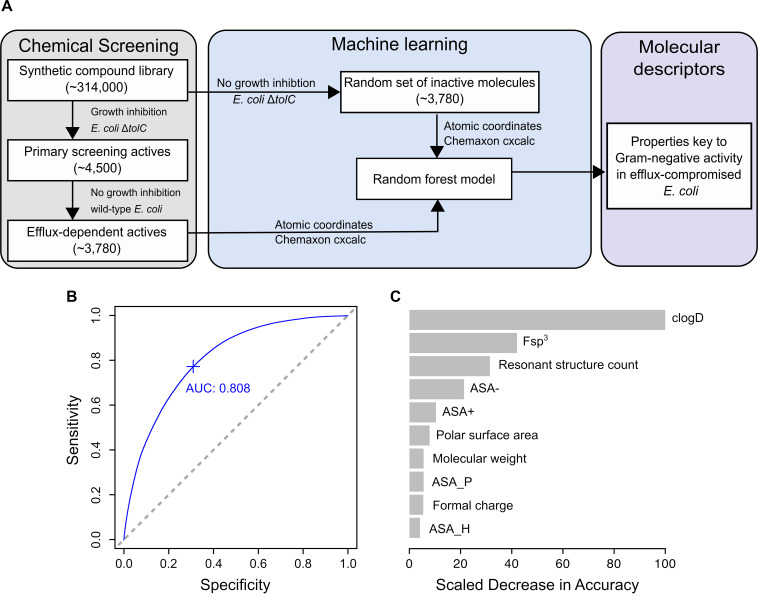
FIG 3.
A machine learning approach identifies key molecular descriptors for Gram-negative activity of efflux-dependent actives. (A) Molecular descriptors for the 3,780 efflux-dependent actives and a random set of 3,780 inactive molecules (no growth inhibition in E. coli BW25113 ΔtolC) from the primary screen were used to train a random forest model to examine descriptors contributing to Gram-negative activity in efflux-compromised E. coli. (B) The area under the curve-receiver operating characteristic curve (AUC-ROC) plot for the random forest model is 0.808, showing a good distinction between efflux-dependent actives and inactive molecules. Sensitivity refers to the true-positive rate of the model, while specificity refers to its false-positive rate. (C) The top 10 molecular descriptors that reduce the model’s accuracy are shown, with clogD, Fsp3 (fraction of sp3 hybridized carbon atoms), and resonant structure count topping the list.