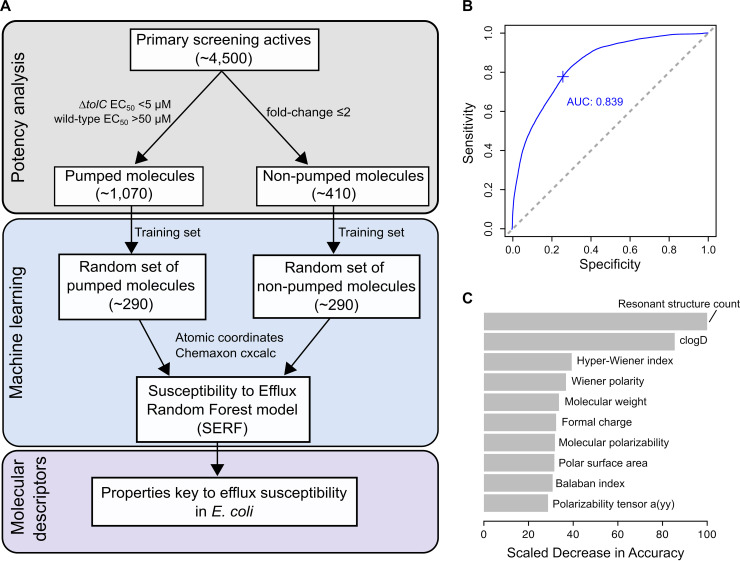
FIG 4.
The Susceptibility to Efflux Random Forest (SERF) model identifies molecular descriptors governing efflux. (A) The pipeline highlights the training set of compounds used to build the SERF model and identify key descriptors that contribute to efflux susceptibility. Based on the shown cutoffs for the 4,500 actives from the primary screen, ∼1,070 actives were pumped molecules and ∼410 actives were nonpumped molecules. For a random set of ∼290 pumped molecules and ∼290 nonpumped molecules, molecular descriptors for each of these compounds were used to train SERF and identify those contributing to efflux susceptibility in E. coli. (B) The area under the curve-receiver operating characteristic curve (AUC-ROC) plot for SERF is 0.839, showing a good distinction between pumped molecules and nonpumped molecules. Sensitivity refers to the true-positive rate of the model, while specificity refers to its false-positive rate. (C) The top 10 molecular descriptors that reduce the model’s accuracy are shown, with resonant structure count, clogD, and hyper-Wiener index accounting for the greatest impact on accuracy. “Polarizability tensor a(yy)” is the principal component of polarizability along the coordinate space a(yy).