Fig. 3.
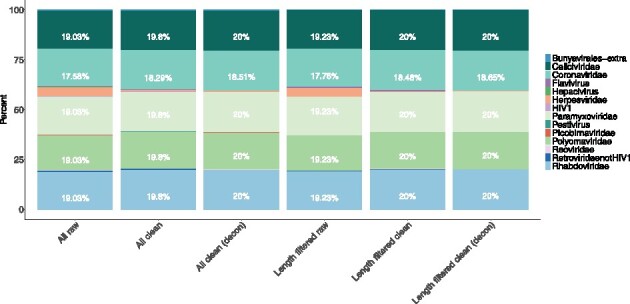
Percentage of mapped reads in the simulated viral metagenomic sample mapping to each of the sub-databases using the kmer-based alignment tool KVIT. The simulated sample contains equal amounts of reads for the five included viral families (Caliciviridae, Coronaviridae, Paramyxoviridae, Polyomaviridae and Rhabdoviridae) and additional contamination reads (phages, human and E.coli). Here, we show the distribution on a viral family level
