Fig. 3.
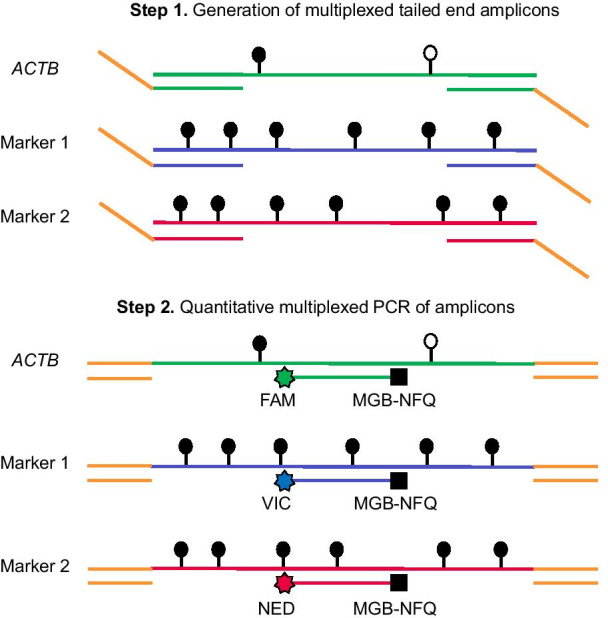
Schema of the TAM-MSP method. The first step of TAM-MSP amplifies two markers and actin control (ACTB) control using a single aliquot of DNA in one well, with primers located within the CpG-rich region of interest for each methylation marker, and therefore includes CpG dinucleotides in its sequence. The 5′ end of the forward and reverse primers for the two methylation markers and ACTB have the same synthetic tails. In the second step of TAM-MSP, primers that are complimentary to the synthetic tails are used, along with marker-specific TaqMan probes, each with one of three indicated fluorescent tags. All three markers are amplified in a single real-time PCR. Methylation in each marker is quantified through interpolation on a historic standard curve and is expressed as percent cumulative methylation. Open circles: unmethylated CpG; closed circles: methylated CpG
