Figure 1.
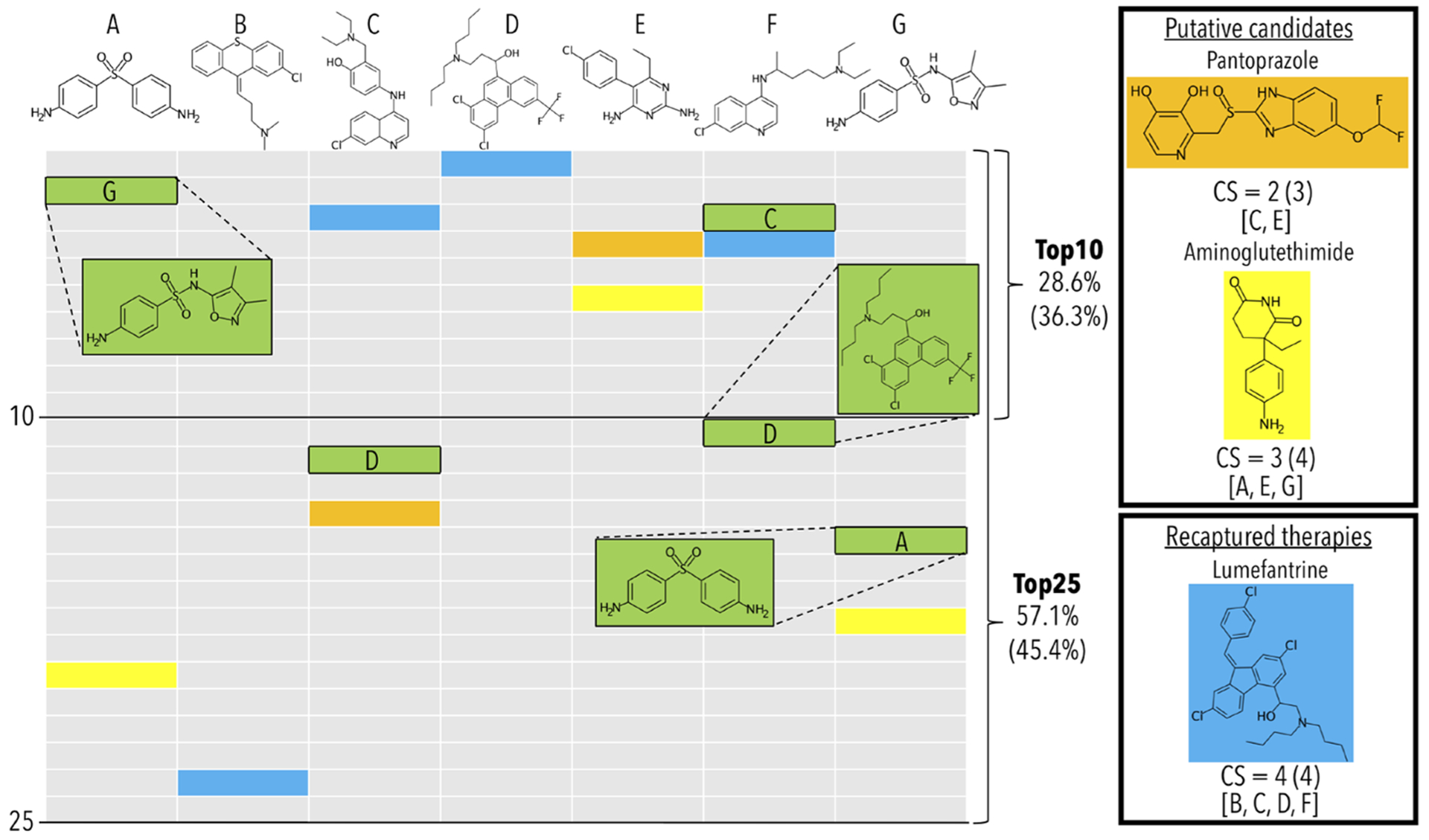
Example of benchmarking and putative therapeutic prediction with the canbenchmark and canpredict modules for malaria (Plasmodium falciparum). A subset of the 22 drugs associated with the indication Malaria Falciparum (MeSH:D016778) are labeled A though G, which from left to right are dapsone, chlorprothixene, amodiaquine, halofantrine, pyrimethamine, chloroquine, and sulfisoxazole. The remaining 15 drugs are excluded for illustrative purposes only. The benchmarking accuracies for the top10 and top25 cutoffs, and the top25 consensus scores (CSs) shown in the figure are based on using only the 7 drug subset (top) and for all 22 drugs (bottom, in parentheses). All drugs and table cells in green are used for calculating benchmarking performance, while the yellow, blue, and orange cells are the predictions highlighted by the canpredict module. The columns represent the ranked order of the most similar drugs/compounds based on root-mean-square-deviations of their proteomic signatures to their respective A through G labeled drug above. The drug–proteome interaction matrix used for this example was created from the interaction scores of 5317 human protein structures from the Protein Data Bank with a library of 2162 approved drugs from DrugBank. The benchmarking method tallies the percent of times another drug associated with the indication is captured within a certain column cutoff rank to a held-out compound (A–G) also associated with the indication. In the example, both drugs A and F, dapsone and chloroquine, recapture another drug associated with the indication within the top10 cutoff, which are sulfisoxazole (G) and amodiaquine (C), respectively. This results in a top10 accuracy of 28.6% (two out of seven). Both amodiaquine (C) and sulfisoxazole (G) recapture another associated drug at the top25 cutoff, which are halofantrine (D) and dapsone (A), respectively. This raises the top25 accuracy to 57.1% (four out of seven). This process is iterated over all indications to calculate global accuracies at each cutoff. The canpredict module utilizes a consensus voting scheme to suggest putative drug repurposing candidates based on the similarity of their proteomic interaction signatures to each known treatment for a disease. A tally is kept of how many times a specific drug is captured within a set cutoff to each known treatment (top25 in this example). Pantoprazole (orange), which has shown antimalarial activity in the literature, falls at rank 14 and 4 for amodiaquine (C) and pyrimethamine (E), respectively, receiving a CS of 2. The aromatase inhibitor aminoglutethimide (yellow) has a CS of 3, which we are suggesting as a novel candidate treatment for malaria. Lumefantrine (blue), a known malaria treatment which in this case was not originally included in the Comparative Toxicogenomics Database drug–indication mapping used by the platform, receives a CS of 4. If lumefantrine was originally included as a treatment, the benchmarking scores would increase to 57.1% and 85.7% at the top10 and top25 cutoffs, respectively, which highlights the importance of drug–indication mapping veracity. The benchmarking module provides insight into how well the given drug–protein interaction scoring method is relating drugs in the context of disease, while the canpredict module suggests putative drug repurposing candidates based on drug– drug similarities.
