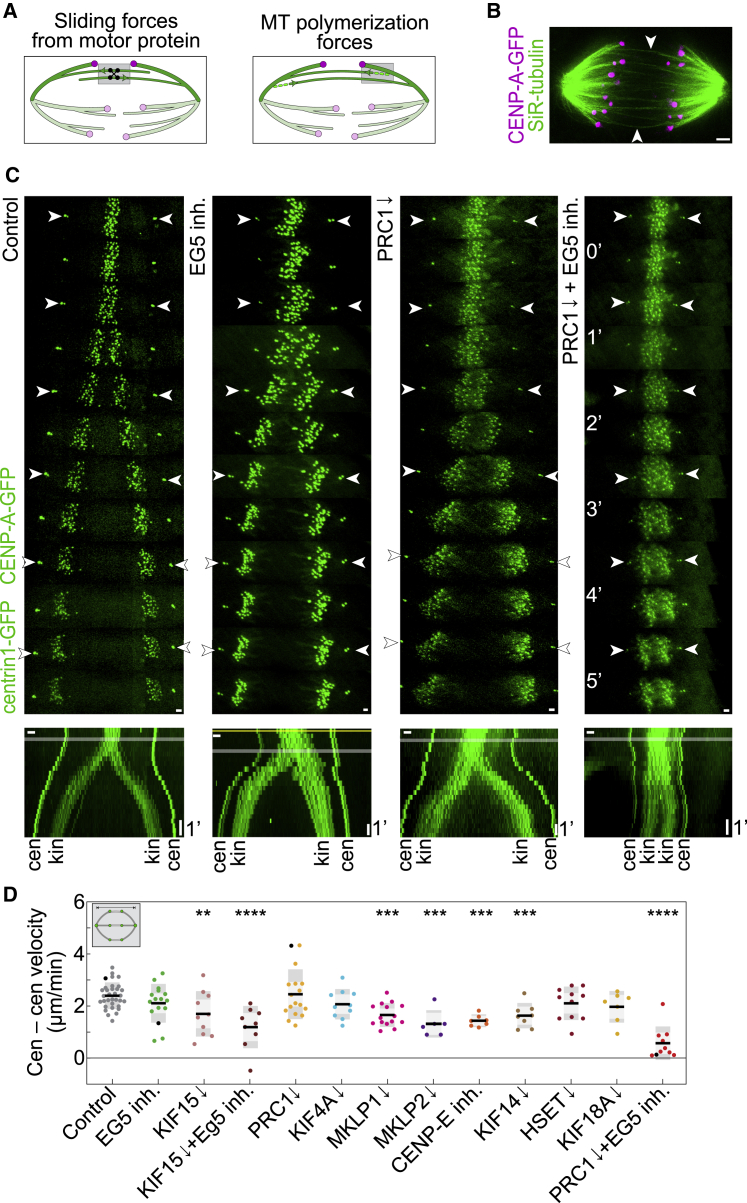
Figure 1.
Depletion of PRC1 and inactivation of EG5 block spindle elongation during anaphase
(A) Two models of spindle elongation.
(B) STED image (single z plane) of an anaphase spindle in a live U2OS cell expressing CENP-A-GFP (magenta). Microtubules are labeled with 100-nM SiR-tubulin (green). Arrowheads point to the spindle midzone where antiparallel microtubules are located.
(C) Live-cell images (top) and corresponding kymographs (bottom) of control, STLC-treated (EG5 inh.), PRC1-siRNA-depleted (indicated by an arrow pointing down) and PRC1-siRNA-depleted 40-μM STLC-treated RPE-1 cells stably expressing CENP-A-GFP and centrin1-GFP. kin, kinetochore; cen, centrosome. White arrowheads indicate centrioles. Horizontal gray lines in the kymographs indicate the onset of anaphase.
(D) Quantification of spindle elongation velocity (see scheme) (n = 34, 16, 10, 9, 17, 10, 15, 6, 7, 8, 11, 7, and 10 cells, from left to right), measured in the period from the 1–3 min after anaphase onset for each treatment. More than three independent experiments for every condition regarding siRNAs or non-targeting treatments, while number of independent experiments regarding STLC treatment is equal to the number of cells. Boxes represent standard deviation (dark gray), 95% confidence interval of the mean (light gray), and mean value (black). Black data dots in every treatment correspond to the measurements from the exemplar cells shown on the time-lapse images and kymographs (C). Statistics: t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001). Time is shown as minutes. Images are the maximum projection of the acquired z stack. Time 0 represents anaphase onset. Horizontal scale bars, 1 μm. Vertical scale bars, 1 min. See also Figure S1.