Figure 2.
Chromosome segregation is compromised after KO of PRC1 and inhibition of EG5
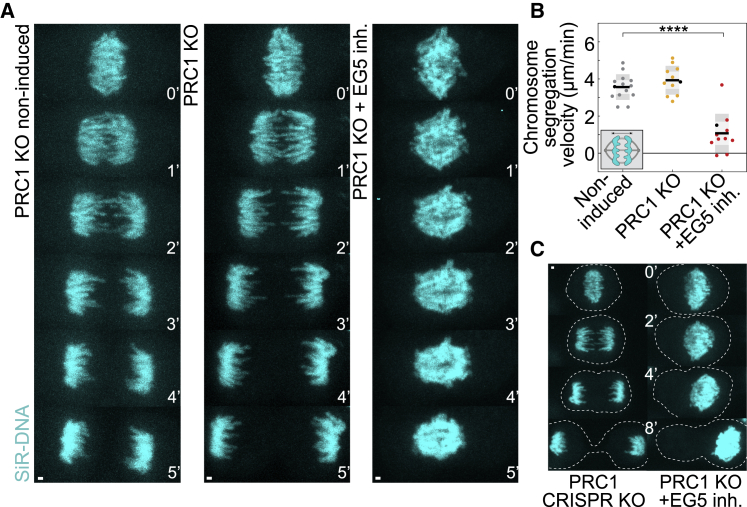
(A) Live images of RPE-1 cells in PRC1 non-induced KO, induced PRC1 CRISPR KO and induced PRC1 KO treated with 40-μM STLC imaged 4 days after doxycycline induction. 100-nM SiR-DNA (cyan) was used for chromosome staining.
(B) Quantification of chromosome segregation velocity (see scheme) in CRISPR experiments (n = 14, 11, and 11, from left to right). Two independent experiments for every condition except PRC1 KO + Eg5 inh. which was done in three independent experiments, while number of independent experiments regarding STLC treatment is equal to the number of cells. Black data dots in every treatment correspond to the measurements from the exemplar cells shown on the time-lapse images and kymographs (A). Statistics: t test (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001).
(C) Live-cell images of induced RPE-1 PRC1 CRISPR KO and induced PRC1 KO treated with 40-μM STLC. Dashed lines designate the cell borders. 100-nM SiR-DNA was used for chromosome staining. Time is shown as minutes. Images are maximum projections of the acquired z stack. Time 0 represents anaphase onset. Scale bars, 1 μm. See also Figure S2.